Article highlight: New insight in how cells regulate gene activity
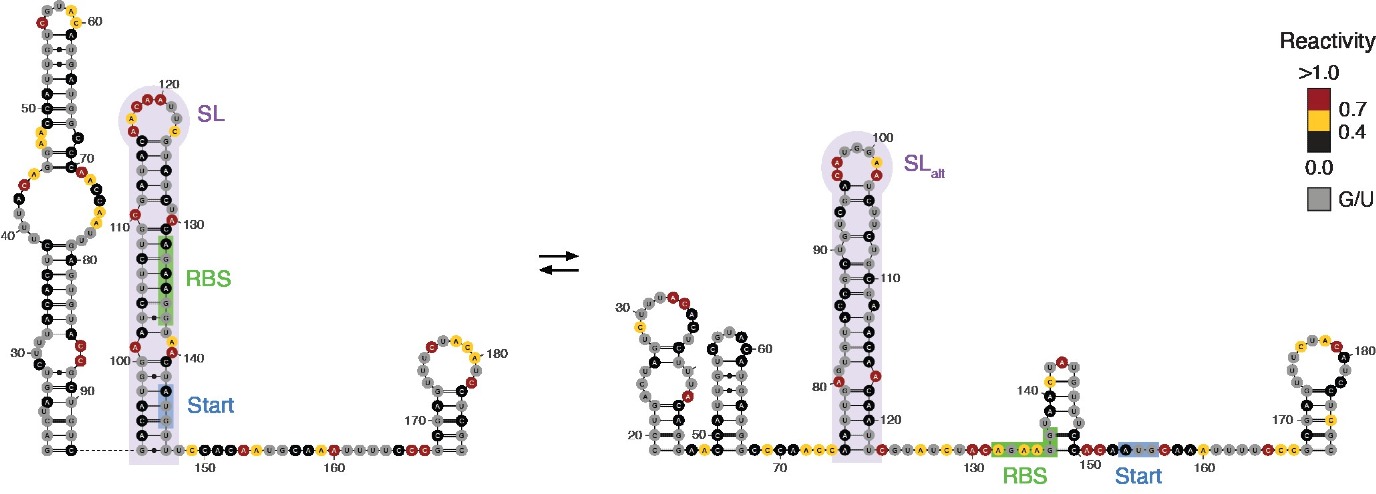
Apart from carrying the information to encode proteins in, RNA molecules can adopt intricate 2D and 3D structures. Specifically, the same RNA molecule can switch between ON and OFF structures, modulating the ability of ribosomes to bind to the RNA and translate it into proteins. A new study, led by University of Groningen molecular biologist Danny Incarnato and authored by postdoctoral researcher Dr. Ivana Borovska, identifies hundreds of such regulatory RNA switches in E.coli bacteria and human cells. It was published in Nature Biotechnology on 25 July.
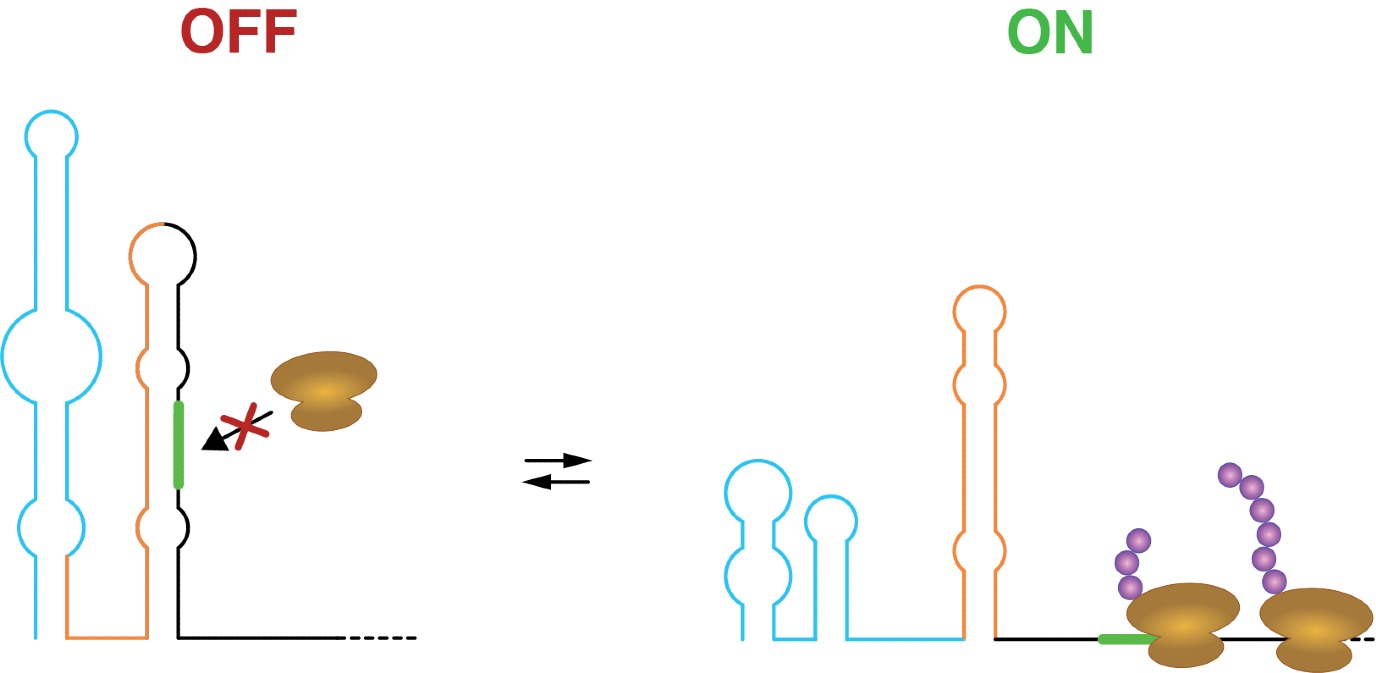
Several years ago, Incarnato developed a method to map the alternative shapes adopted by RNA molecules in living cells. Using this method, he found regions of RNA capable of shapeshifting between two different structures, each with its own effect. Incarnato: ‘The ability of RNA to switch between alternative structures usually implies some sort of regulation, similar to an ON-OFF switch.’
New treatments
Incarnato’s team has now used this method to study the complexity of RNA structures in living cells. Moreover, they developed a new tool that leverages evolutionary information to identify functional RNA structural switches with high accuracy. They used the tool to uncover hundreds of them. One example is a switch that reacts to temperature and helps bacteria to respond to cold stress. Incarnato: ‘Identifying a significant number of switches is a first step. The next step is to find ways to influence their functioning.’ For example, small molecules could be designed to modulate these switches, which could ultimately lead to new treatments for diseases.
The current research, which has taken more than three years to complete, builds on some six years of fundamental research into the detection of 2D RNA shapes. Incarnato: ‘This is revolutionary, something the whole field has been seeking for years.’
Reference: Ivana Borovská, Danny Incarnato et al. Identification of conserved RNA regulatory switches in living cells using RNA secondary structure ensemble mapping and covariation analysis. Nature Biotechnology, 25 July 2025.
More news
-
06 January 2026
Getting to grips with the workhorses of our body
