Multiscale method development
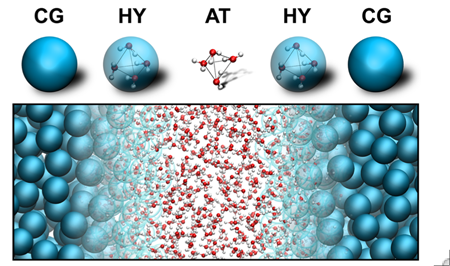
To take full benefit of the power of computational microscopy, the resolution of the computational microscope should be tuneable. In particular, techniques that can couple the sampling speed gained with coarse-grain models to the accuracy inherent of all-atom models are needed.
We pioneer the use of such multiscale techniques, e.g., we develop efficient algorithms to transform configurations between different levels of resolution (‘backmapping’), we introduced a novel method to perform hybrid simulations using virtual sites, and we explore the coupling of atomistic force fields to MARTINI in adaptive resolution simulations.
[1] T.A. Wassenaar, K. Pluhackova, R.A. Böckmann, S.J. Marrink, D.P. Tieleman. Going backward:A flexible geometric approach to reverse transformation from coarse grained to atomistic models. JCTC, 10:676-690, 2014
[2] A.J. Rzepiela, M. Louhivuori, C. Peter, S.J. Marrink. Hybrid simulations: combining atomistic and coarse-grained force fields using virtual sites. Phys. Chem. Chem. Phys., 13:10437-10448, 2011
[3] T.A. Wassenaar, H.I. Ingólfsson, M. Prieß, S.J. Marrink, L.V. Schäfer. Mixing Martini: electrostatic coupling in hybrid atomistic – coarse-grained biomolecular simulations. J. Phys. Chem. B, 117:3516–3530, 2013
[4] J. Zavadlav, M.N. Melo, S.J. Marrink, M. Praprotnik. Adaptive resolution simulation of an atomistic protein in MARTINI water. J. Chem. Phys., 140:054114, 2014
[5] N. Goga, M.N. Melo, A.J. Rzepiela, A.H. De Vries, A. Hadar, S.J. Marrink, H.J.C. Berendsen. Benchmark of schemes for multiscale molecular dynamics simulations. JCTC, 11:1389–1398, 2015.
| Last modified: | 24 June 2015 11.43 p.m. |
