High throughput membrane builder
Research in the MD group often involves (biological) membranes of many different compositions. We're developing several tools to efficiently build, simulate and analyse membranes.
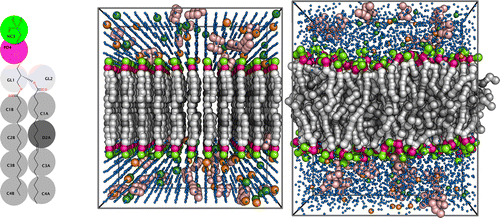
Building custom membranes and protein-membrane systems can be performed using the Insane script [1], which allows simple and versatile system setup. This does not just aid MD users to setup their specific systems, but also allows the streamlining of 'lipidomics' simulations, where huge numbers of different membranes can be studied with a few simple commands.
One of the scientific problems such tools can be applied to is the aggregation of membrane proteins and/or peptides. By performing large numbers of simulations of how e.g. dimers associate one can find the most probable binding sites.[2] The high-throughput treatment is necessary in this case to gain sufficient statistical significance.
[1] T.A. Wassenaar, H.I. Ingólfsson, R.A. Böckmann, D.P. Tieleman, S.J. Marrink. Computational lipidomics with insane: a versatile tool for generating custom membranes for molecular simulations. JCTC, 11:2144–2155, 2015. doi: 10.1021/acs.jctc.5b00209
[2] T.A. Wassenaar, K. Pluhackova, A. Moussatova, D. Sengupta, S.J. Marrink, D.P. Tieleman, R.A. Böckmann. High-throughput simulations of dimer and trimer assembly of membrane proteins. The DAFT approach. JCTC, 11:2278–2291, 2015.
| Last modified: | 12 January 2016 4.16 p.m. |
