A cricket that tracks antibiotic resistance
In just a few weeks, a team of 13 students from the University of Groningen will travel to Paris. From 28-31 October, at the Grand Jamboree of the iGEM competition, they will present ‘CRIKIT’, a portable kit that makes detecting antibiotic resistance in bacteria faster, cheaper and more accessible to everyone.
Science LinX | Text: Myrna Kooij | Images: CRIKIT
Antibiotic resistance is a growing global threat to public health. The World Health Organization recently warned that this is making simple bacterial infections, such as pneumonia, more difficult to treat. When bacteria are repeatedly exposed to antibiotics, they adapt, rendering once-effective drugs useless. Detecting resistance can currently take days and often requires expensive lab equipment that is not widely available. For this, the University of Groningen’s iGEM team has found a solution.
We thought of the Cas13 protein as a cricket that jumps onto genes and checks: ‘Are you the one I’m looking for?’ So it just jumps around like a cricket.-- Mihai Frăcea
Cutting RNA
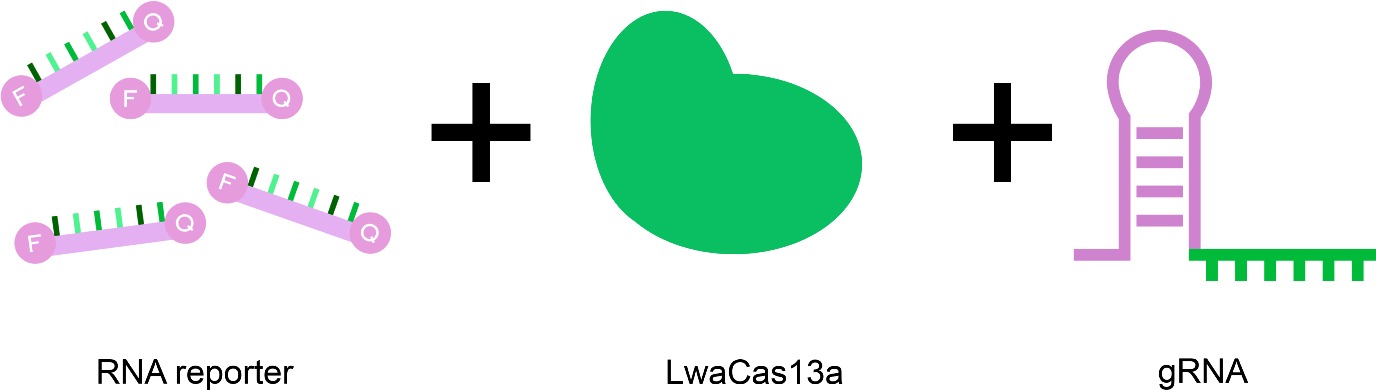
The CRIKIT works with CRISPR Cas13-technology, which targets the RNA of antibiotic-resistant bacteria. Team member Mihai Frăcea simplifies: “You can imagine Cas13 as a pair of scissors with a GPS. The Cas13 enzyme is the scissors; it does the cutting, and it also gives us a fluorescent signal.” This light signal reports when antibiotic resistance is detected.
But the scissors need directions to know where to cut. For that, a guide RNA (gRNA), also called a ‘spacer’, comes in. Frăcea continues: “The spacer is the GPS. It orients, it tells the scissors where to go, what to look for.”
By using different types of spacers, CRIKIT can detect various resistance genes. “The kit is basically unchanged, except for the spacers. That's the only part that you should change based on local problems,” Frăcea explains. This is because resistance genes can vary across different regions. Frăcea gives an example: “What infections might be in the Netherlands might not be the same for Brazil and might not be the same for Southeast Asia.” Frăcea adds: “So you have to adapt your test very easily for those conditions, and our test can actually do that.”
Speed is key
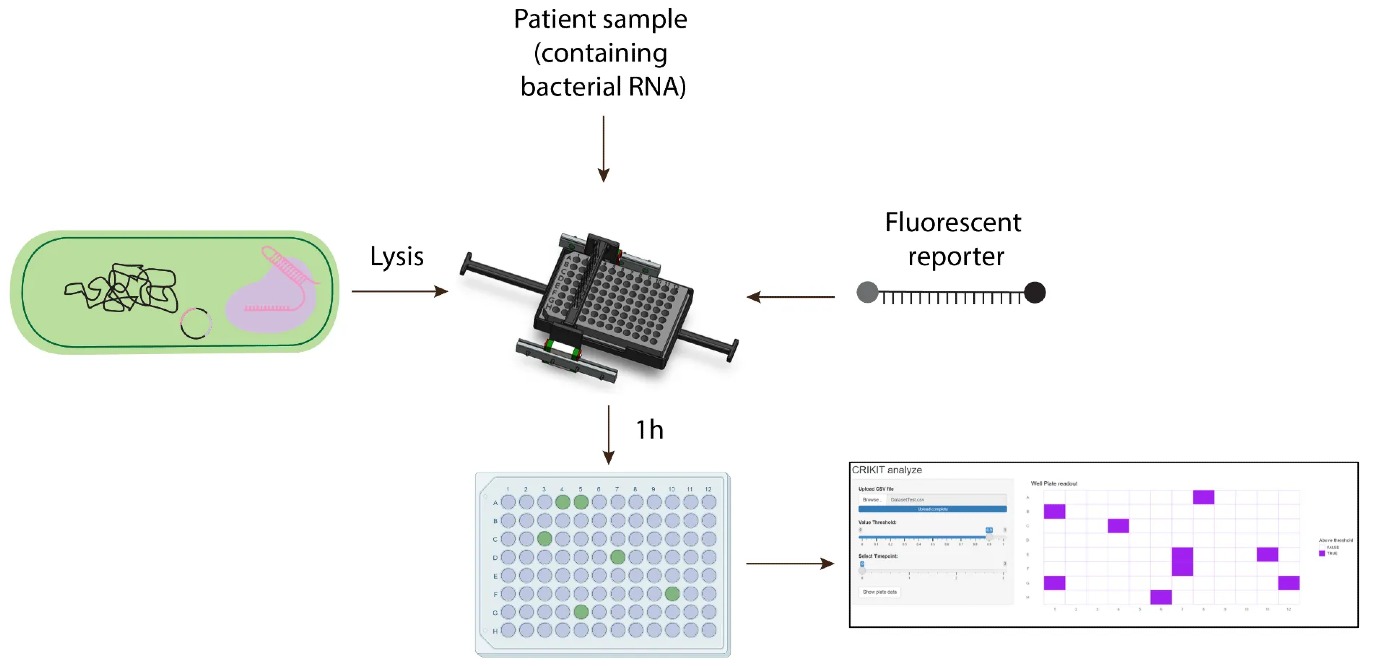
In the hospital, time is of the essence for patients in critical conditions. Frăcea: “We've actually talked to a medic, and he had a case when a person died of pneumonia in two days. And I didn't think you could die of pneumonia in 2025. But it was a very resistant pathogen.” Frăcea highlights that current tests available to doctors are slow. “You have to test extremely fast, under 24 hours if possible,” says Frăcea. “Sometimes you don’t even have 24 hours.” That’s why the detection kit’s key element is speed. Their current estimation is that their kit takes about 2 hours, but they’re working hard to make it even faster.
An affordable and portable kit
Next to time-efficiency, the team aims for their design to be available for everyone. “Detecting fluorescence in most cases is very expensive,” Frăcea explains. He estimates plate readers can cost up to a million euros. “It's a huge box and it's very heavy. So you cannot really take it and move it around, and it's not really field-deployable.”
To fix that challenge, the team decided to build their own version of a plate reader. Frăcea proudly credits team member Dilara Yigit for the design of CRIKIT’s hardware. “It's just a box in which you put your samples, and it's 3D printed, so pretty cheap,” says Frăcea. “It's basically a plate reader that instead of costing a million, it costs around maybe a few hundred euros. And you can even reuse it.”


The plate reader’s design is small yet loaded with multiple gadgets, using some simple LED lights, photo-sensors and filters to measure the fluorescence. In addition, the box also comes with Bluetooth and Wi-Fi to send the readings to a computer, where the team’s own software analyzes the data and shares an overview of the genes of the patient’s bacterial infection that show resistance to antibiotics. “Instead of using expensive machinery that is very heavy and difficult to move, you just have this little black box in which you put the samples,” Frăcea illustrates.
From healthcare to agriculture
While the team’s current focus is on antibiotic resistance, CRIKIT’s core design (detecting genetic material) is employable for other purposes outside of healthcare. “Our kit could very easily be adapted for agriculture,” Frăcea suggests. “We can easily detect, for example, infections or bacteria that affect crops. Because they also have this problem of antibiotic resistance in agriculture, both in terms of crops, but also in terms of cows and feedstock.” The team has already approached a potato breeding company for a collaboration.



Team effort
The CRIKIT team started working together on their project already in February. “It was just beautiful to see how everybody brings their own input into the project,” Frăcea reminisces. “I would define our brainstorming meetings as a lot of aha moments.” Frăcea's biggest lesson? "We can do much more than we thought we could do at this level of master's students, which is very encouraging. And that we can actually get results with enough planning and constant work.”
Would you like to follow the team’s journey? Go follow the CRIKIT team on Instagram or LinkedIn and check out their entire project on their iGEM wiki page.

The International Genetically Engineered Machine (iGEM) Competition is a yearly event where students from all over the world are challenged to solve a real-world problem by genetically engineering a microorganism. Every year, a team from the University of Groningen participates.
More news
-
15 September 2025
Successful visit to the UG by Rector of Institut Teknologi Bandung

