New publication on the Nuclear Pore Complex in Nature Communications
The latest research of the Onck group, “Kap-centric Nsp1-mediated nuclear transport at full amino acid resolution,” was just published in Nature Communications.
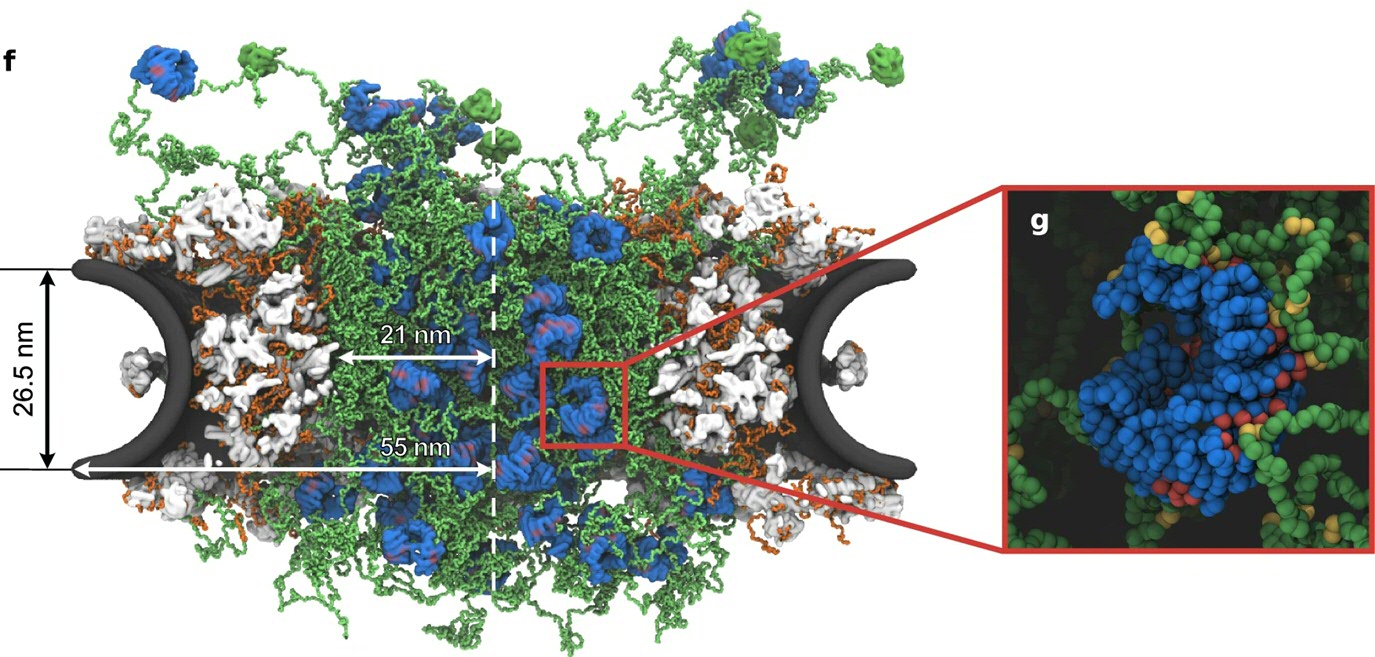
In this work, Onck and colleagues present the first full amino acid–resolution model of nuclear pore complex (NPC) transport, enabling large-scale simulations of selective transport through the pore. Their study reveals how the dynamic FG-Nup meshwork—centered around the bimodal architecture of Nsp1—regulates both passive and active transport. By explicitly incorporating nuclear transport receptors (Kap95), they uncover how Kaps reinforce the permeability barrier while navigating the pore through transient, fluctuation-driven pathways.
As Prof. Onck notes, this paper brings together over 15 years of preparatory work, starting from an early project under the Bonus Incentive Scheme (with Van der Giessen and Veenhoff) that set out to model nuclear transport at unprecedented molecular detail—capturing all amino acids inside the megadalton transport channel in a single framework. Thanks to the outstanding contributions of our PhD researchers Ghavami, Mishra, Jafarinia, De Vries and Dekker, this long-standing challenge has now been met, yielding not only new mechanistic insight but also striking visualizations and a compelling simulation movie.
A milestone for computational biophysics and nuclear transport research.
Read the paper in Nature Communications: https://www.nature.com/articles/s41467-025-66329-z
More news
-
15 September 2025
Successful visit to the UG by Rector of Institut Teknologi Bandung
