A closer look at the Huntington-protein
Solid-state Nuclear Magnetic Resonance (ssNMR) enables the study of the structure of the mutated Huntington-protein – the cause of Huntington’s Disease. This has been reported by the group of professor Van der Wel from the Zernike Institute (at the time professor in Pittsburgh) and a research group led by professor De Paëpe in Grenoble in France in the scientific journal Journal of the American Chemical Society. ‘If we can better understand that structure, we might be able to develop smarter drugs that target the cause of the disease, rather than just treating the symptoms.’
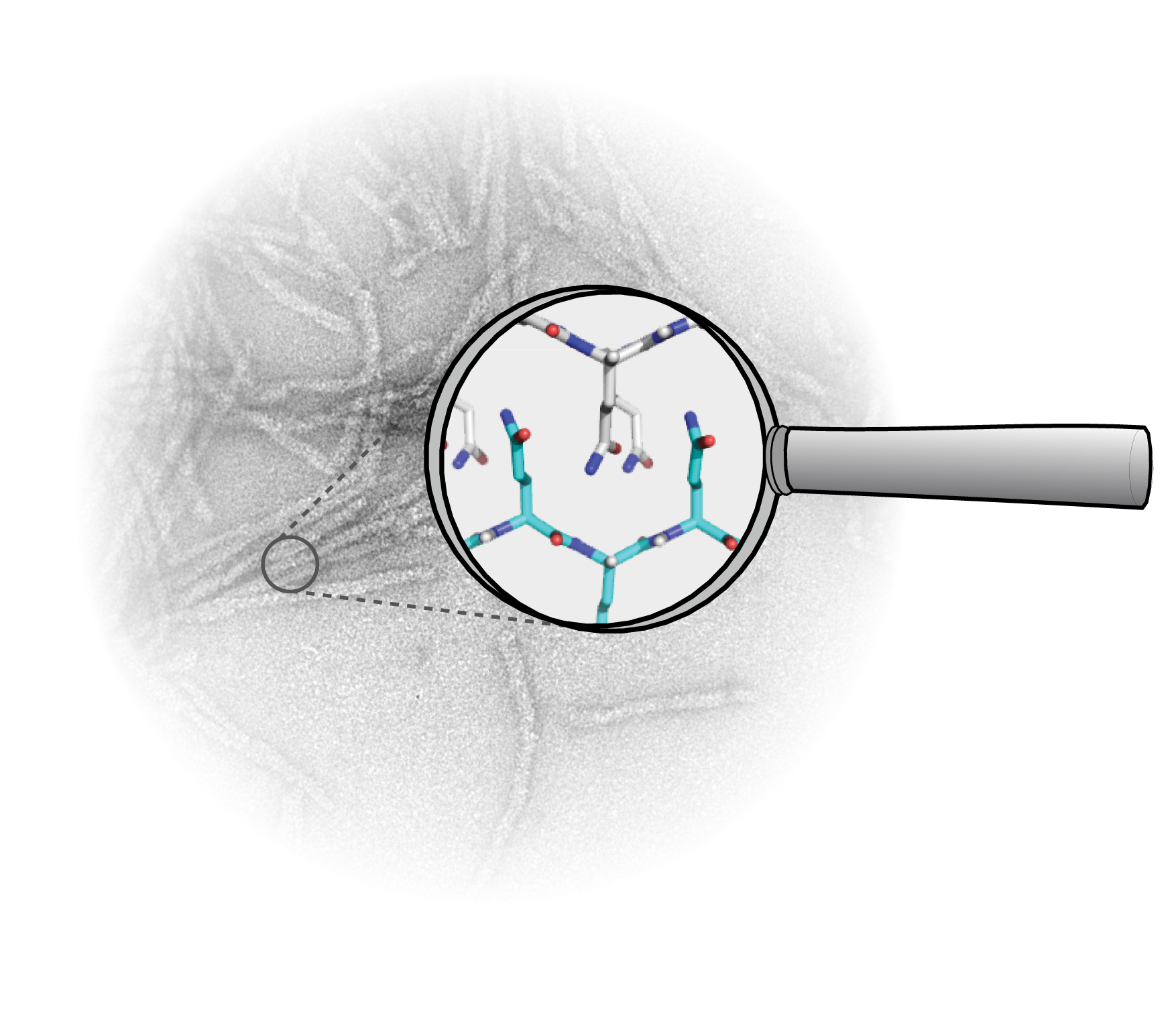
Huntington’s Disease is a so-called neurodegenerative disease, such as Parkinson’s Disease and Alzheimer’s Disease. Many neuronal cells in the central nervous system die over time in neurodegenerative diseases. The outcome, amongst other things, is deterioration of the motor skills and the memory and eventually leads to a premature death.
Protein aggregates
In contrast to many other neurodegenerative diseases, the cause of Huntington’s Disease is relatively clear and predictable: patients have a special kind of mutation in their DNA that is responsible for the before mentioned huntingtin-protein. The mutation leads to misfolding of this protein and consequently the formation of protein aggregates, a type of cluster of proteins. Such protein clusters are also seen in other neurodegenerative diseases. It is not known if and how these protein clusters contribute to the death of the neurons, partially because the structure of the protein is not known yet.
In the heart of the protein
It is not easy to image the protein. That is not possible yet with commonly used techniques based on X-ray diffraction and high-resolution electron microscopy. ‘If you look at the structure of the clustered protein, you can understand why this is such a challenge,’ explains Van der Wel. ‘It forms kind of a long wire from thousands of proteins, of which no crystals can be made. In addition, these wires do not dissolve easily and are disordered and dynamic.’ Two years ago, the Van der Wel group managed for the first time to look into the heart of the protein, in the mutated part. This was possible via a special form of spectroscopy, based on the detection of nuclear magnetic resonance, which is also used in MRI scanners. ‘However, that was only possible when we made the protein ourselves in the laboratory with extra 13C-atoms in it. We needed these atoms to get a good look at the protein. 13C-atoms are rare in nature: naturally only one in a hundred carbon-atoms is 13C. Thus, that is also the situation you find in proteins of natural sources, which makes it difficult to study protein clusters from humans and animals with our technique.’
Natural proteins
This indicates the importance of this new research. ‘We have now shown that we can carefully look at the protein without adding 13C. This can make it possible in the future to look at huntingtin-proteins from organisms or living cells to see if the protein we study in the lab has the exact same structure. That is an essential next step in understanding the molecular origin of the disease.’ Hopefully such information can play a role in the development of both drugs and signaling molecules that recognize and detect the misfolded proteins (for the benefit of diagnosis and prognosis).
Other proteins
A better understanding of Huntington’s Disease might also help in understanding (and targeting) other neurodegenerative diseases. ‘Unfortunately, it is not easy to apply this technique directly on other proteins. To also look at proteins from Parkinson’s Disease and Alzheimer’s Disease more research has to be done. But our French colleagues will definitely continue with developing this research method.’
Smith A.N., Märker K., Piretra T., Boatz J.C., Matlahov I., Kodali R., Hediger S., van der Wel P.C.A., De Paëpe G. Structural fingerprinting of protein aggregates by DNP-enhanced solid-state NMR at natural isotopic abundance. J Am Chem Soc. 2018;140(44):14576-80. https://pubs.acs.org/doi/10.1021/jacs.8b09002
More news
-
15 September 2025
Successful visit to the UG by Rector of Institut Teknologi Bandung
