Biophysics
In this work we study fundamental processes in the cell in order to elucidate the biophysical mechanisms that are responsible for transport of biomolecules through the nuclear pore complex, the transmission of mechanical forces through the cytoskeleton, protein aggregation in neurodegenerative diseases and membrane fusion through conformational dynamics of surface proteins.
1. The Nuclear Pore Complex: gate keepers of the nucleus
Although it has long been assumed that proteins only inherit their function from folding into a specific structure, there is now increasing evidence that intrinsically disordered proteins play an important role in many biological functions, such as nuclear transport. Transport of molecules in and out of the nucleus of eukaryotic cells is controlled by large protein complexes, called ‘nuclear pore complexes’ (NPCs), that are embedded in the nuclear membrane. The actual transport is mediated by intrinsically disordered proteins, FG-nucleoporins (FG-nups), forming a low-density cloud of flexible filaments that line the core region of the NPC. In this project we explore how these FG-nups mediate transport through coarse-grained molecular dynamics simulations and explore how this is affected in case of the neurodegenerative disease ALS.
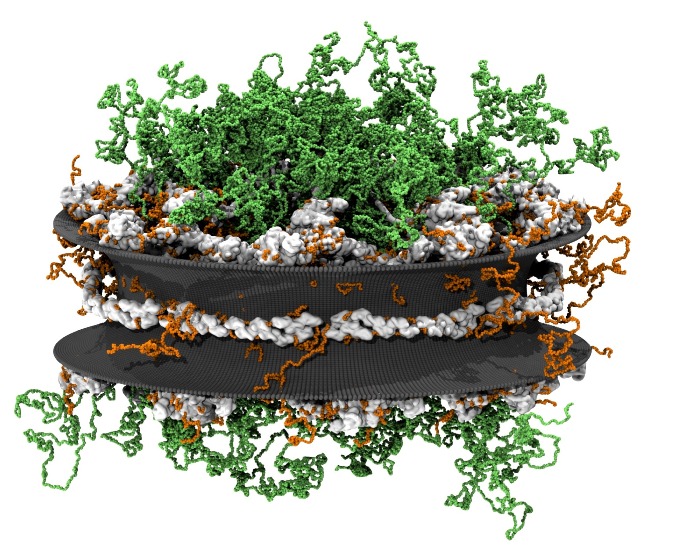
PhD students: M. Dekker, A. Mishra, A. Ghavami, H. de Vries
In collaboration with: L. Veenhoff (ERIBA, Groningen); C. Dekker (Delft University of Technology).
Publications (selection)
-
Dekker, M., Van der Giessen, E., & Onck, P. R. (2023). Phase separation of intrinsically disordered FG-Nups is driven by highly dynamic FG motifs. Proceedings of the National Academy of Sciences of the United States of America, 120(25), Article e2221804120. https://doi.org/10.1073/pnas.2221804120
-
Klughammer, N., Barth, A., Dekker, M., Fragasso, A., Onck, P. R., & Dekker, C. (2023). Diameter Dependence of Transport through Nuclear Pore Complex Mimics Studied Using Optical Nanopores. eLife, 12, Article eLife.87174.1. Advance online publication. https://doi.org/10.7554/eLife.87174.1
-
Fragasso, A., de Vries, H. W., Andersson, J., van der Sluis, E. O., van der Giessen, E., Onck*, P. R., & Dekker*, C. (2022). Transport receptor occupancy in nuclear pore complex mimics. Nano Research, vol. 15, 9689–9703 (2022), https://doi.org/(...)07/s12274-022-4647-1
-
Fragasso, A., de Vries, H. W., Andersson, J., van der Sluis, E. O., van der Giessen, E., Dahlin, A., Onck*, P. R., & Dekker*, C. (2021). A designer FG-Nup that reconstitutes the selective transport barrier of the nuclear pore complex. Nature Communications, 12(1), 2010. https://doi.org/(...)8/s41467-021-22293-y
-
Hoogenboom, B. W., Hough, L. E., Lemke, E. A., Lim, R. Y. H., Onck, P. R., & Zilman, A. (2021). Physics of the nuclear pore complex: Theory, modeling and experiment. Physics Reports , Vol. 921, 25 July 2021, Pages 1-53. https://doi.org/(...).physrep.2021.03.003
-
Rempel, I. L., Popken, P., Ghavami, A., Mishra, A., Hapsari, R. A., Wolters, A. H. G., ... Veenhoff, L. M. (2020). Flexible and Extended Linker Domains Support Efficient Targeting of Heh2 to the Inner Nuclear Membrane. Structure, 28(2), 185-195.e5. https://doi.org/(...)16/j.str.2019.11.003
-
Rempel, I. L., Crane, M. M., Thaller, D. J., Mishra, A., Jansen, D. P. M., Janssens, G., ... Veenhoff, L. M. (2019). Age-dependent deterioration of nuclear pore assembly in mitotic cells decreases transport dynamics. eLife, 8, [e48186]. DOI: 10.7554/eLife.48186
-
Mishra, A., Sipma, W., Veenhoff, L. M., Van der Giessen, E., & Onck, P. R. (2019). The Effect of FG-Nup Phosphorylation on NPC Selectivity: A One-Bead-Per-Amino-Acid Molecular Dynamics Study. International Journal of Molecular Sciences, 20(3), [596]. DOI: 10.3390/ijms20030596
-
Ghavami, A., Van der Giessen, E., & Onck, P. R. (2018). Sol-gel transition in solutions of FG-Nups of the nuclear pore complex. Extreme Mechanics Letters, 22, 36-41. DOI: 10.1016/j.eml.2018.04.006
-
Ketterer, P., Ananth, A. N., Laman Trip, D. S., Mishra, A., Bertosin, E., Ganji, M., ... Dekker, C. (2018). DNA origami scaffold for studying intrinsically disordered proteins of the nuclear pore complex. Nature Communications, 9(1), [902]. DOI: 10.1038/s41467-018-03313-w
-
Ananth, A. N., Mishra, A., Frey, S., Dwarkasing, A., Versloot, R., van der Giessen, E., ... Onck*, P.R. Dekker*, C. (2018). Spatial structure of disordered proteins dictates conductance and selectivity in Nuclear Pore Complex mimics. eLife, 7. DOI: 10.7554/eLife.31510, *Corresponding authors.
-
Fuertes, G., Banterle, N., Ruff, K. M., Chowdhury, A., Mercadante, D., Koehler, C., ... Lemke, E. A. (2017). Decoupling of size and shape fluctuations in heteropolymeric sequences reconciles discrepancies in SAXS vs. FRET measurements. PNAS, 114(31), E6342-E6351. DOI: 10.1073/pnas.1704692114
-
Ghavami, A., van der Giessen, E., & Onck, P. R. (2016). Energetics of Transport through the Nuclear Pore Complex. PLoS ONE, 11 (2), 1-13. [e0148876]. DOI: 10.1371/journal.pone.0148876
-
Popken, P., Ghavami, A., Onck, P. R., Poolman, B., & Veenhoff, L. M. (2015). Size-dependent leak of soluble and membrane proteins through the yeast nuclear pore complex. Molecular Biology of the Cell, 26 (7), 1386-1394. DOI: 10.1091/mbc.E14-07-1175
-
Ghavami, A., Veenhoff, L. M., van der Giessen, E., & Onck, P. R. (2014). Probing the Disordered Domain of the Nuclear Pore Complex through Coarse-Grained Molecular Dynamics Simulations. Biophysical Journal, 107 (6), 1393-1402. DOI: 10.1016/j.bpj.2014.07.060
-
Ghavami, A., van der Giessen, E., & Onck, P. R. (2013). Coarse-Grained Potentials for Local Interactions in Unfolded Proteins. Journal of Chemical Theory and Computation, 9 (1), 432-440. DOI: 10.1021/ct300684j
2. Biopolymer Networks
Polymer networks are ubiquitous in living matter (e.g., cytoskeleton and extracellular matrix) and are upcoming in the form of engineered fiber networks. Irrespective of their nature, the mechanical properties of these networks depend on the properties of the filament and crosslinks, as well as on the architecture. We use a combination of a FEM designed for filaments and crosslinks along with Monte Carlo techniques to study the elastic and time-dependent properties of these networks.
PhD students: A. Boerma, G. Zagar; Postdoc: T. van Dillen
In collaboration with: S. Papanikolaou (University of West Virginia) for Monte Carlo modeling.
Publications (selection)
-
Shakibi, S., Onck, P. R., Van, E., & van der Giessen, E. (2023). Extracellular proteoglycans: A multiscale computational study. Biophysical Journal, 122(3 Supplement 1), 420a-421a. https://doi.org/10.1016/j.bpj.2022.11.2279
-
Shakibi, S., Onck, P. R., & Van der Giessen, E. (2023). A One-Bead-Per-Saccharide (1BPS) Model for Glycosaminoglycans. Journal of Chemical Theory and Computation, 19(16), 5491–5502. https://doi.org/10.1021/acs.jctc.3c00238
-
Zagar, G., Onck, P. R., & van der Giessen, E. (2015). Two Fundamental Mechanisms Govern the Stiffening of Cross-linked Networks. Biophysical Journal, 108 (6), 1470-1479. DOI: 10.1016/j.bpj.2015.02.015
-
Zagar, G., Onck, P. R., & Van der Giessen, E. (2011). Elasticity of Rigidly Cross-Linked Networks of Athermal Filaments. Macromolecules, 44 (17), 7026-7033. DOI: 10.1021/ma201257v
-
Zagar, G., Onck, P. R., & Giessen, E. V. D. (2009). Mechanics of Biophysical Networks with Flexible Cross-links. Biophysical Journal, 96 (3), 122a-123a. DOI: 10.1016/j.bpj.2008.12.544
-
van Dillen, T., Onck, P. R., & Van der Giessen, E. (2008). Models for stiffening in cross-linked biopolymer networks: A comparative study . Journal of the Mechanics and Physics of Solids, 56 (6), 2240-2264. DOI: 10.1016/j.jmps.2008.01.007
-
Huisman, E. M., van Dillen, T., Onck, P. R., & Van der Giessen, E. (2007). Three-dimensional cross-linked F-actin networks: Relation between network architecture and mechanical behavior. Physical Review Letters, 99 (20), [208103]. DOI: 10.1103/PhysRevLett.99.208103
-
Onck, P. R., Koeman, T., Van Dillen, T., & Van der Giessen, E. (2005). Alternative Explanation of Stiffening in Cross-Linked Semiflexible Networks. Physical Review Letters, 95 (17), [178102]. DOI: 10.1103/PhysRevLett.95.178102
3. Influenza viral fusion
All types of influenza rely on the replication of viruses inside host cells. Replication requires that the virus is able to inject its RNA into the cell, which in turn needs fusion of the viral membrane with the cell membrane. It is known that the protein hemagglutinin acts as a catalyst for this fusion process, but how it does so at the right speed is a long-standing question. Using GROMACS on massively parallel computers, we perform molecular dynamics simulations of hemagglutinin at atomic resolution with the aim to better understand the gymnastics that drive membrane fusion. Identification of a critical step in these conformational changes that is robust for all types of hemagglutinin opens the avenue to a universal physics-based anti-viral drug.
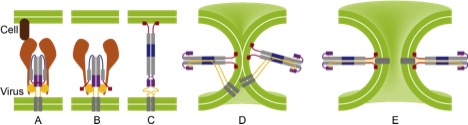
PhD student: S. Boonstra
In collaboration with: A.M. van Oijen (University of Wollongong) and PhD student J.S. Blijleven for experimental background and support.
Publications
-
Boonstra, S., Blijleven, J. S., Roos, W. H., Onck, P. R., van der Giessen, E., & van Oijen, A. M. (2018). Hemagglutinin-Mediated Membrane Fusion: A Biophysical Perspective. Annual Review of Biophysics, 47. DOI: 10.1146/annurev-biophys-070317-033018
-
Boonstra, S., Onck, P. R., & Van der Giessen, E. (2017). Computation of Hemagglutinin Free Energy Difference by the Confinement Method. The Journal of Physical Chemistry. B, 121(50), 11292-11303. DOI: 10.1021/acs.jpcb.7b09699
-
Blijleven, J. S., Boonstra, S., Onck, P. R., van der Giessen, E., & van Oijen, A. M. (2016). Mechanisms of influenza viral membrane fusion. Seminars in Cell & Developmental Biology. DOI: 10.1016/j.semcdb.2016.07.007
-
Boonstra, S., Onck, P. R., & van der Giessen, E. (2016). CHARMM TIP3P Water Model Suppresses Peptide Folding by Solvating the Unfolded State. The Journal of Physical Chemistry. B: 120 (15), 3692-3698. DOI: 10.1021/acs.jpcb.6b01316
4. Protein aggregation in neuro-degenerative diseases
Most neurodegenerative diseases, such as ALS, Alzheimer’s, Parkinson’s and Huntington’s disease, rarely occur at a young age, but its incidence increases dramatically in older people, reaching 10–25% at the age of eighty. These age-related diseases are caused by the aggregation of intrinsically disordered proteins into stable filamentous amyloids that are toxic. The underlying molecular mechanisms of self-assembly and aggregation has been subject of intense debate, but a coherent picture is still lacking. The aim of this project is to identify the specific molecular mechanism behind this protein aggregation, which, if successful, will enhance our understanding of amyloidogenic protein aggregation and might open up new avenues for drug targeting.
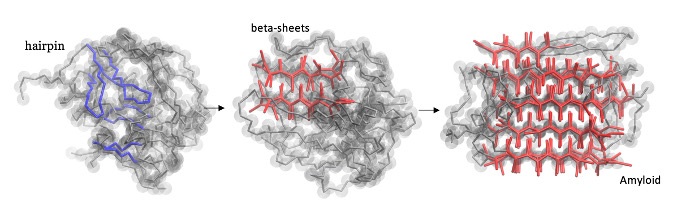
PhD students: V. Adupa, M. van der Klok, H. Jafarinia, Postdoc: M. Driver
Publications
-
Heesink, G., Marseille, M. J., Fakhree, M. A. A., Driver, M. D., van Leijenhorst-Groener, K. A., Onck, P. R., Blum, C., & Claessens, M. M. A. E. (2023). Exploring Intra- and Inter-Regional Interactions in the IDP α-Synuclein Using smFRET and MD Simulations. Biomacromolecules, 24(8), 3680–3688. https://doi.org/10.1021/acs.biomac.3c00404
-
Jafarinia, H, Van der Giessen, E & Onck, PR 2022, 'Molecular basis of C9orf72 poly-PR interference with the β-karyopherin family of nuclear transport receptors', Scientific Reports, vol. 12, no. 1, 21324. https://doi.org/10.1038/s41598-022-25732-y
-
Van der Klok, M., Dekker, M., Van der Giessen, E., & Onck, P.R. (2021). A 4BPA Coarse-Grained Molecular Dynamics Study on the Aggregation of Polyglutamine. Biophysical Journal, 120(3 Suppl. 1), 28A-28A. https://doi.org/(...)16/j.bpj.2020.11.428
-
Driver, M. D., & Onck, P. R. (2021). RNA-Protein Interactions in Molecular Life and Health: A Computational Approach. Biophysical Journal, 120(3), 81A-81A. https://doi.org/(...)16/j.bpj.2020.11.702
-
Pappas, C. G., Liu, B., Marić, I., Ottelé, J., Kiani, A., van der Klok, M. L., Onck, P. R. , & Otto, S. (2021). Two Sides of the Same Coin: Emergence of Foldamers and Self-Replicators from Dynamic Combinatorial Libraries. Journal of the American Chemical Society , 143(19), 7388–7393. https://doi.org/10.1021/jacs.1c00788
-
Jafarinia, H., van der Giessen, E., & Onck, P. R. (2020). Phase Separation of Toxic Dipeptide Repeat Proteins Related to C9orf72 ALS/FTD. Biophysical Journal, 119(4), 843-851. https://doi.org/(...)16/j.bpj.2020.07.005.
