Research Highlights
Prof. dr. Dick Janssen
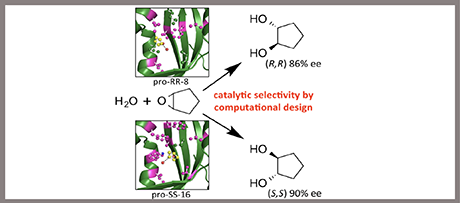
Enzyme engineering by computational library design and in silico screening
Directed evolution has emerged as a powerful tool for tailoring enzyme properties to specific biotechnological targets. Its main bottleneck is the large amount of laboratory work necessary for discovering improved variants in the vast sequence space that is accessible even by only a few mutations. To reduce and streamline the required laboratory work, the Janssen group investigated the use of computational design screening of mutant enzymes. This yielded a workflow (FRESCO, framework for rapid enzyme stabilization by computation) that involves in silico design and screening of mutant libraries, mainly using comparative energy calculations and high-throughput molecular dynamics simulations, which were carried out in collaboration with the Molecular Dynamics group (Marrink). For experimental verification, the workflow was applied to enzymes that catalyze reactions of biotechnological relevance, including epoxide hydrolase, dehalogenases, and peptide amidase. This produced robust enzyme variants that exhibit a large increase in thermostability (ΔTm,app +15-35ºC) and high cosolvent compatibility. The resulting stabilized enzymes proved to be good templates for further mutagenesis aimed at selectivity engineering which is again supported by computational design and screening using molecular dynamics.
| Last modified: | 04 July 2017 10.12 a.m. |
