Protein Sequencing

The large-scale study of the proteins produced by an organism, proteomics, is crucial to understand cellular processes underlining diseases and to identify specific indicators linked to diseases in a biological sample, also known as biomarkers.
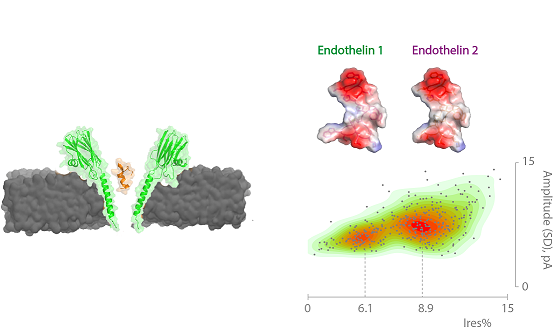
FraC nanopores for protein sequencing
Publications
Willems K, Ruić D, Biesemans A, Galenkamp N, Van Dorpe, P, Maglia G. Engineering and Modeling the Electrophoretic Trapping of a Single Protein Inside a Nanopore. ACS Nano. (2019) doi.org/10.1021/acsnano.8b09137
Huang G, Voet A, and Maglia G. FraC Nanopores with Adjustable Diameter Identify the Mass of Opposite-Charge Peptides with 44 Dalton Resolution. Nature Commun. (2019) Feb 19. doi: 10.1038/s41467-019-08761-6
|
BOX 1: DNA SEQUENCING Building on this work, a commercial device that sequences DNA, the Oxford Nanopore Technologies (ONT) MinION, is being tested in clinics. The device has a low capital cost, is by far the most portable DNA sequencer available, and can produce data in real-time. Because it can perform long reads, it has numerous prospective applications including the ability of improving genome sequence assemblies and resolution of repeat-rich regions. Arrays of thousands of nanopores allow high-throughput analysis. However, owing to the multiple occupancy of nucleobases inside the nanopore used, the raw output in the MinION is generated not by individual bases but by 5-nucleotide stretches known as k-mers. |
| Last modified: | 12 December 2019 2.32 p.m. |
