Nanopore Sensing
Biomarkers are molecules that offer a measurable indicator of some biological state or condition. The presence of biomarkers in biological samples is used to monitor pathogenic processes. Using nanopore is advantageous because they do not require chemical labelling or complex optics thus allowing easy integration in low-cost, portable and wearable devices.
In collaboration with Oxford Nanopore Technology, we are currently developing a nanopore based technology for high-throughput sensing of biomarkers.
Metabolite Recognition with Protein Adaptors
Metabolites are small molecules that are products of metabolism and are an important class of biomarkers.
Proteins can be integrated inside a nanopore and the binding of ligands can be observed by the specific modulation of the ionic current. Metabolites can be identified directly from picoliter blood samples, sweat and other biological liquids.
We are testing the incorporation of many proteins that bind to a variety of metabolites.
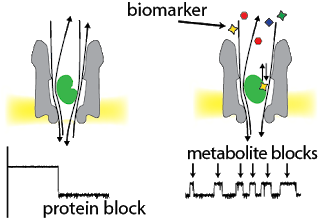
Nanofilters for protein biomarkers
Many biomarkers are proteins. Protein biomarkers are recognised by their specific ionic current signature (e.g. thrombin in green and GFP in blue) and quantified by the frequency of the blockades.
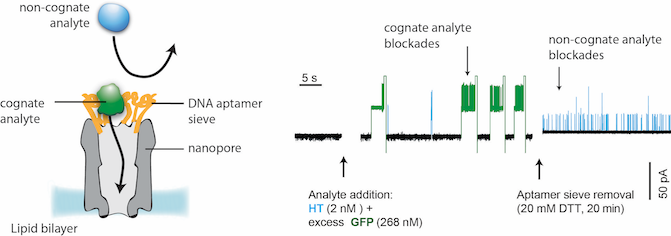
One of the main challenges is to detect low abundance proteins in a biological sample. We are investigating several methods to selectively recognize a protein biomarker. In one example below, a layer of DNA aptamers was chemically attached to the nanopore to allow selective capture of cognate analyses.
Publications
-Eurekalert - Sciencelinx - PhysOrg - Science News - Science Daily - Canada News - LongRoom - GEN - R&D - Infosurhoy - Bionity - Health Innovations - Bioanalysis - Kennislink - Chemistry World - LabMedica
Van Meervelt V, Soskine M, Singh S, Schuurman-Wolters G, Wijma HJ, Poolman B, and Maglia G. Real-time conformational changes and controlled orientation of native proteins inside a protein nanoreactor. J Am Chem Soc. (2017) Dec 27;139(51):18640-18646. doi: 10.1021/jacs.7b10106. PDF
Willems K, Van Meervelt V, Wloka C, and Maglia G. Single-molecule nanopore enzymology. Philos Trans R Soc Lond B Biol Sci. (2017) Aug 5;372(1726). pii: 20160230. DOI: 10.1098/rstb.2016.0230. PDF
Soskine M, Biesemans A, Maglia G. Single-Molecule Analyte Recognition with ClyA Nanopores Equipped with Internal Protein Adaptors. J Am Chem Soc. 6;137(17):5793-7 (2015). DOI: 10.1021/jacs.5b01520.
Van Meervelt V, Soskine M, and Maglia G. Detection of two isomeric binding configurations in a protein-aptamer complex with a biological nanopore. ACS Nano Dec 23;8(12):12826-35 (2014). DOI: 10.1021/nn506077e.
Further readings
Nanopore sensing (2016)
| Last modified: | 04 February 2020 10.59 a.m. |
