DNA analysis
Single-stranded nucleic acid molecules can be driven through a nanometer size pore by an applied electric field. As each molecule occupies the pore, a characteristic blockade of ionic current is produced. Information about length, composition, structure, dynamic motion, and sequence of the molecule can be deduced from modulations of the current blockade.
Nanopores are now used to sequence or map DNA at low cost and high speed. We are studying the fundamental properties of DNA in the confinement of the nano pores. We have developed biological nano pores that are capable of analyzynig double strand DNA molecules at physiological ionic strengths.
DNA analysis with FraC nanopores
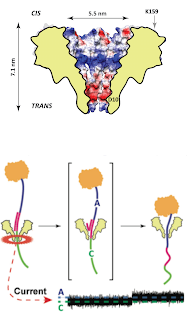
We have characterised a new nanopore FraC for DNA analysis. FraC has a narrow constriction (~ 1.2 nm) that is ideal for DNA analysis.
We found that homoplymeric strands show a different signal when they are immobilised inside the nanopore.
At low applied potential only single stranded DNA can traverse the nanopore, while at high applied potential, the alpha helical transmembrane part of the nanopore opens to let double stranded DNA through.
Therefore, FraC might be used in DNA sequencing applications or employed in the analysis of double stranded DNA
Mechanism of DNA translocation across nanopores
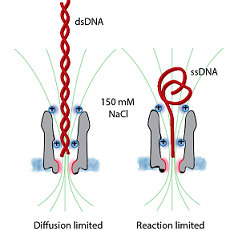
The translcoation of DNA across ClyA nanopores required the precise engineering of the nanopore internal surface charge, and only when initiated from the wider side of the nanopore.
We proposed a mechanism where the engineered positive charges are important to align the DNA in order to overcome the entropic and electrostatic barriers for DNA translocation through the narrow constriction.
Our results also suggest a physical model where the capture of double-stranded DNA is diffusion-limited while the capture of single-stranded DNA is reaction-limited.
Publications
Stoddart D, Franceschini L, Heron A, Bayley H, Maglia G. DNA stretching and optimization of nucleobase recognition in enzymatic nanopore sequencing. Nanotech. (2015) Feb 27;26(8):084002. DOI
Franceschini, L., Mikhailova, E., Bayley, H., and Maglia, G. Nucleobase recognition at alkaline pH and apparent pKa of single DNA bases immobilised within a biological nanopore. ChemComm 1;48(10):1520-2 (2012). DOI
Restrepo, M.R., Mikhailova, E., Bayley, H., and Maglia, G. Controlled translocation of individual DNA molecules through protein nanopores with engineered molecular brakes. Nano Lett 11(2):746-50 (2011). DOI
| Last modified: | 08 October 2019 09.19 a.m. |
