Synthetische data versnellen onderzoek naar gist
Wetenschappers bestuderen de groei en celdeling van bakkersgist vaak met behulp van time-lapse microscopie. Systemen op basis van kunstmatige intelligentie zijn daarbij zeer nuttig, zij kunnen veranderingen in individuele cellen herkennen en volgen door de serie beelden. Maar voor het trainen van zo’n KI systeem is een grote hoeveelheid nauwkeurig beschreven data nodig, en het kost maanden om die te produceren. Onderzoekers van de RUG hebben nu laten zien dat met behulp van synthetische data het trainen slechts enkele dagen kost. Het nieuwe systeem werkt net zo goed als de beste beschikbare neurale netwerken voor de analyse van gistcellen.
Bakkersgist is een van de best bestudeerde organismen ter wereld. Gistcellen dienen als model voor fundamentele processen in complexe cellen, zoals die van mensen. Maar er is toch nog veel dat we niet over gistcellen weten, legt assistent hoogleraar computationele biologie Andreas Milias-Argeitis uit: ‘Een belangrijke vraag die op antwoord wacht is hoe gistcellen hun groei controleren tijdens de celcyclus. Anders gezegd: hoe stemmen ze de toename in biomassa af op de replicatie van DNA, de productie van essentiële bouwstenen en het hele proces van celdeling?’

Training
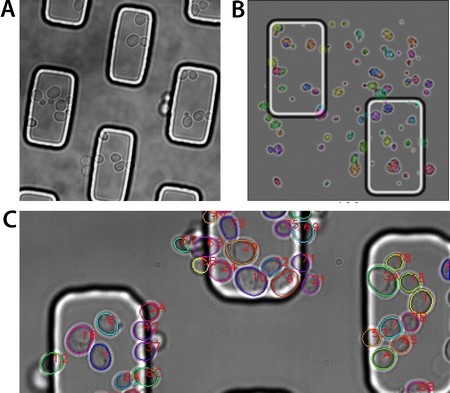
De belangstelling van Milias-Argeitis ligt op het snijvlak van biologie en computeranalyse, en hij bestudeert deze vragen in grote hoeveelheden gegevens van time-lapse microscopie. ‘We observeren hoe honderden cellen groeien en delen gedurende vele generaties, en kijken daarbij naar specifieke processen. We kunnen de cellen ook verstoren, door bijvoorbeeld bepaalde genetische paden uit of aan te schakelen met behulp van licht, een techniek die optogenetica heet.’ Door individuele cellen te volgen in een tijdreeks van beelden is het mogelijk om te zien hoe een verstoring het uiterlijk van de cel verandert. Maar dit soort experimenten produceert ontzettend veel gegevens, dus is er een vorm van geautomatiseerde beeldanalyse nodig. Convolutionele neurale netwerken (CNN’s) zijn daar geschikt voor, maar deze systemen moeten worden getraind om de cellen te herkennen.
‘Dat gebeurt door een CNN microscopische beelden aan te bieden waarin iemand de omtrek van duizenden cellen heeft gemarkeerd’, legt Milias-Argeitis uit. Die annotatie moet perfect zijn, wat een nogal tijdrovende klus is. ‘Een student doet er al snel maanden over om zo’n set trainingsdata te produceren.’ En als de onderzoeksvraag verandert, moet het trainingsprogramma weer opnieuw beginnen. Een snellere manier om zo’n CNN te trainen zou het onderzoek daarom flink versnellen.

Synthetische data
Dus toen het Centrum voor Informatietechnologie (CIT) van de RUG een oproep deed voor het indienen van onderzoeksvoorstellen in data-wetenschap stelde Milias-Argeitis de vraag of er een sneller trainingsmethode mogelijk was. Zijn voorstel werd geaccepteerd door het CIT en hij kreeg hulp van datawetenschapper Herbert Kruitbosch. Zij begonnen te werken aan een idee dat Kruitbosch had gesuggereerd: een trainingsmethode gebaseerd op synthetische data. Milias-Argeitis: ‘Het gebruik van synthetische data om KI systemen te trainen is zeker in de biologie nog niet vaak toepast. Maar Herbert had zeer veel ervaring met beeldverwerking en hij heeft een goed oog voor kenmerkende structuren in cellen. En dat terwijl hij weinig afweet van gist – of misschien juist daardoor.’
Kruitbosch produceerde een dataset met op gist lijkende vormen en speelde vervolgens met allerlei instellingen, zoals voor vervorming van cellen, hun grootte en de ruis in het beeld. Deze synthetische dataset is vervolgens gebruikt om een CNN (van het type Mask R-CNN) te trainen voor beeldverwerking. Het getrainde systeem is daarna getest op echte data van gistcellen. ‘En het werkte direct verrassend goed. Ik kon het zelfs niet geloven toen ik de eerste resultaten zag.’ De prestaties van het nieuwe systeem zijn uiteindelijk vergeleken met die van een van de beste CNN’s die was getraind met echte data. Beide bleken even goed te presteren. Maar het grote voordeel van de synthetische data is dat een set voor de training in een dag is te produceren. Bovendien kost het hoogstens een paar dagen om het systeem nieuwe taken te leren. Ten slotte is het erg gebruiksvriendelijk: het kost maar een paar uur om er mee om te leren gaan.
Gebeurtenissen
‘Dit alles betekent dat wij onze experimenten nu veel sneller kunnen ontwerpen en uitvoeren’, zegt Milias-Argeitis. ‘Omdat het getrainde CNN werkt zonder hulp van de gebruiker is het zelfs mogelijk om het in te zetten voor real-time dataverwerking, gericht op veranderingen in celgedrag tijdens een experiment. We kunnen nu bijvoorbeeld een experiment uitvoeren onder de microscoop waarbij we de locatie van individuele cellen bepalen en hun reactie volgen op een optogenetische verstoring, om vervolgens op basis van de reactie de optogenetische input per cel af te stemmen.’
Het project met het CIT is inmiddels afgerond en de resultaten zijn op 10 december 2021 gepubliceerd in het tijdschrift Bioinformatics. Alle software en algoritmes zijn beschikbaar gesteld in een vrij toegankelijke database, zodat iedereen er gebruik van kan maken. ‘We werken nu aan de verdere ontwikkeling van ons KI systeem, bijvoorbeeld om het te leren hoe het specifieke gebeurtenissen tijdens de celdeling kan herkennen, of mutante cellen met een afwijkende vorm. We verwachten dat we het meeste werk zelf kunnen doen, maar gelukkig is Herbert beschikbaar als adviseur.’
Referentie: Herbert Kruitbosch, Yasmin Mzayek, Sara Omlor, Paolo Guerra and Andreas Milias-Argeitis: A convolutional neural network for segmentation of yeast cells without manual training annotations. Bioinformatics, 10 december 2021
Software en algortimes zijn beschikbaar in deze publieke database
Meer nieuws
-
10 maart 2026
Microplastics als een boemerang
-
17 februari 2026
De lange zoektocht naar nieuwe fysica

