
New Key Publication in Cell: The long-term genetic stability and individual specificity of the human gut microbiome
Highlights
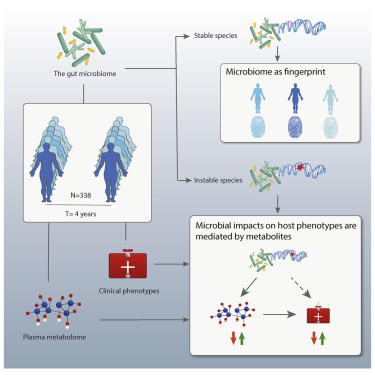
- Gut microbial genetic stability differs across species
- Gut microbial composition with higher baseline diversity is more stable over time
- Individual-specific, temporally stable microbial profiles can fingerprint the host
- The microbial impact on host health is mediated by plasma metabolites
By following up the gut microbiome, 51 human phenotypes and plasma levels of 1,183 metabolites in 338 individuals after 4 years, we characterize microbial stability and variation in relation to host physiology. Using these individual-specific and temporally stable microbial profiles, including bacterial SNPs and structural variations, we develop a microbial fingerprinting method that shows up to 85% accuracy in classifying metagenomic samples taken 4 years apart. Application of our fingerprinting method to the independent HMP cohort results in 95% accuracy for samples taken 1 year apart. We further observe temporal changes in the abundance of multiple bacterial species, metabolic pathways, and structural variation, as well as strain replacement. We report 190 longitudinal microbial associations with host phenotypes and 519 associations with plasma metabolites. These associations are enriched for cardiometabolic traits, vitamin B, and uremic toxins. Finally, mediation analysis suggests that the gut microbiome may influence cardiometabolic health through its metabolites.
By:
- Lianmin Chen
- Daoming Wang
- Sanzhima Garmaeva
- Alexander Kurilshikov
- Arnau Vich Vila
- Ranko Gacesa
- Trishla Sinha
- Lifelines Cohort Study
- Eran Segal
- Rinse K. Weersma
- Cisca Wijmenga
- Alexandra Zhernakova
- Jingyuan Fu
Read more : Published:April 09, 2021DOI:https://doi.org/10.1016/j.cell.2021.03.024
UMCG press release: https://umcgresearch.org/w/the-gut-microbiome-stable-or-unstable-over-time-
More news
-
15 September 2025
Successful visit to the UG by Rector of Institut Teknologi Bandung
