Research Scheffers Group
Bacterial Cell Biology
Pathogenic bacteria that are resistant to most or all classes of existing antibiotics are rapidly emerging and are the cause of many problematic infections in hospitals. Notorious examples include Methicillin Resistant Staphylococcus aureus (MRSA), multi-drug resistant Mycobacterium tuberculosis and Acinetobacter baumannii. Alarmingly, nearly all antibiotics currently in the drug development pipelines are variations on old themes, making it likely that multidrug resistant bacteria will readily acquire resistance against these not-so-novel drugs. Therefore there is an urgent need for new drug targets. In this respect, bacterial cell division is one of the most promising targets for the development of new antibacterial drugs. We are exploring cell division as a target to combat a plant pathogen - Xanthomonas citri subsp citri - that infects citrus trees. This work we do in collaboration with Dr. Henrique Ferreira, from Rio Claro, Brazil and dr. P. Deuss at ENTEG. Some of the techniques we use are explained in a VIDEO that you can see here.
In collaboration with Prof. P. Olinga (GRIP) and Prof. A. Casini (Univ. of Cardiff, UK) we study gold compounds for antibacterial properties and modes of action.
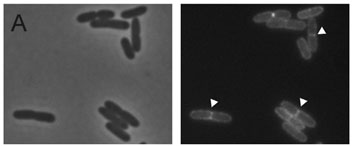
Finally, we are very interested in the shape and division of bacteria. We study fundamental aspects of cell wall synthesis in Bacillus subtilis, a Gram-positive model bacterium. The main component of the cell wall is peptidoglycan, and this giant molecule is built up of little bricks that are all exactly the same. The question we ask is how it is possible to use this same building block to make differently shaped pieces of peptidoglycan - some relatively flat, some curved and some at almost straight angles. Part of the answer is in how the building blocks are linked together during the construction of the wall, and how this process is coordinated. We use fluorescence microscopy to determine the location of all the proteins involved in the process as well as the building blocks. We combine this with genetic or chemical approaches to disturb the process to see what happens in controlled situations. Next to microscopy, we use biochemical methods to determine whether or not the proteins involved cooperate in large protein machineries, as is presumed by most scientists in the field. In 2015, we were the first to show that this is the case for proteins involved in cell division. In recent years we have shown that the machinery that builds the wall is coordinated by the availability of building blocks, and that the modifications to these building blocks are indeed different in different places in the cell. We have also shown, together with Prof. Marc Bramkamp, from Kiel, Germany, (link: https://www.bramkamplab.org/) that the physical properties of the cell membrane, in which the synthesis machineries are embedded, are critical for the movement of these machineries and determine how fast they can construct a cell wall.
| Laatst gewijzigd: | 15 december 2021 17:03 |
