Peroxisome-vacuole membrane contact sites
Master student project, 2020
Aim
The aim of this project is to identify proteins that play a role in vacuole-peroxisome membrane contact sites (VAPCONS) in the yeast Hansenula polymorpha.
Background
All processes that take place inside the cell need to be coordinated and in balance. To achieve this, eukaryotic cells have cell organelles, where functionally specialized processes take place. Until recently, it was believed that inter-organelle communication and exchange of molecules are achieved with molecular diffusion through the cytosol and vesicular transport. Now, we know that membrane contact sites (MCSs) play crucial roles in these processes.
MCSs are regions where two organelle membranes are in close proximity. At MCSs membranes tether, but do not fuse. MSCs have a defined lipid and protein composition, which support MCS function. These include tether proteins and proteins that are important for the function of the MCS (for instance transporter proteins). Recently, an MCS between peroxisomes and vacuoles was described in the yeast Hansenula polymorpha, named VAPCONS. It was shown that the peroxisomal membrane protein Pex3 accumulates in patches at this contact site. Moreover, artificial overproduction of Pex3 can stimulate its formation. Pex3 complex pull down and mass spectrometry analysis was performed, which resulted in a list of protein candidates present in VAPCONS. To confirm which of these candidates are real VAPCONS components, we will investigate their localization with fluorescence microscopy. Also, we will analyze their function in strains that lack or overproduce these proteins.
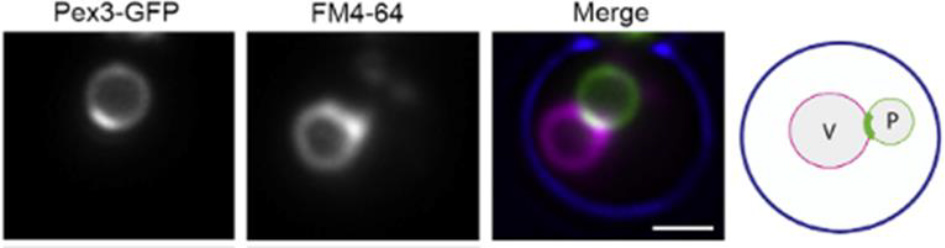
Approach
• Construction of strains in which VAPCON protein candidates are fused with a fluorescent protein (e.g. GFP).
• Construction of strains in which genes encoding promising VAPCON proteins are deleted or overexpressed.
• Validation of prepared strains with DNA sequencing. Analysis of protein levels by Western blotting
• Co-localization studies of VAPCON candidates and Pex3 protein with fluorescence microscopy.
• Co-localization studies of VAPCON candidates and vacuoles with fluorescence microscopy.
• Analysis of the effect of deletion /overexpression of VAPCON candidate genes.
Techniques
Bacteria and yeast handling, in silico experimental design with Snap gene and /or Clone manager, PCR, molecular cloning with restriction and ligation, agarose electrophoresis, bacteria and yeast transformation, fluorescence microscopy, image processing with ImageJ, SDS page and Western blotting.
Literature
Scorrano, L., De Matteis, M. A., Emr, S., Giordano, F., Hajnóczky, G., Kornmann, B., ... & Rizzuto, R. (2019). Coming together to define membrane contact sites. Nature communications, 10(1), 1-11.
Wu, H., de Boer, R., Krikken, A. M., Akşit, A., Yuan, W., & van der Klei, I. J. (2019). Peroxisome development in yeast is associated with the formation of Pex3-dependent peroxisome-vacuole contact sites. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research, 1866(3), 349-359.
Contact Daily supervisor: Tjasa Kosir, Ph.D. student
Supervisor: Ida van der Klei, prof. dr. i.j.van.der.klei@rug.nl
Molecular Cell Biology Faculty of Science and Engineering University of Groningen, Netherlands
| Last modified: | 25 August 2020 1.55 p.m. |
