Research Highlights
Prof.dr. Bert Poolman
Recent breakthroughs by the Poolman group:
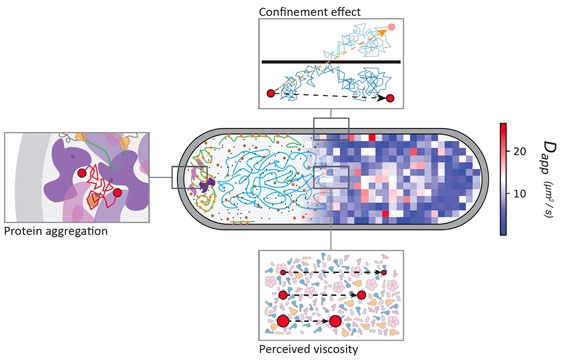
Protein diffusion in living cells
We have developed a sensitive single-molecule displacement method for analyzing simulation-based reconstructed diffusion of proteins in small compartments, which we have successfully applied to investigate the relationship between the dynamics of proteins in the cell pole regions and aging of bacterial cells[1]. We use the method to analyze the diffusion of a wide range of proteins in the Escherichia coli cytoplasm to demonstrate that diffusion is fastest in the nucleoid region of the cell and that the newly formed pole of dividing cells exhibits a faster diffusion than the old one; the latter is due to the accumulation of aggregated or damaged proteins.
| Last modified: | 20 October 2023 2.12 p.m. |
