Scientists identify potential drug target in the new coronavirus

Scientists from the University of Groningen, together with colleagues from the IIMCB in Warsaw (Poland) and the Leiden UMC (also in the Netherlands), have identified putative drug targets in the RNA of the SARS-CoV-2 coronavirus that is causing the current pandemic. Their results, which still have to undergo peer review, show which portions of the genetic material would be promising drug targets. This information can guide the search for a drug that will combat the coronavirus.
The manuscript with the results was uploaded on Monday 15 June to the bioRxiv preprint server. Preprint servers are used by scientists to provide rapid access to their colleagues. In coronavirus research, this is important as normal publishing procedures can take many months. However, as the manuscript has not yet been reviewed by experts, it might be subjected to corrections and improvements at this stage. The authors have also submitted their work for a rigorous peer-review. Currently, new information about coronavirus research is quickly picked up by the media, and this includes preprints.
The study was coordinated by University of Groningen assistant professor Danny Incarnato. His lab is involved in research on the structure of RNA, the genetic material in viruses like SARS-CoV-2. The RNA contains the genetic code for the production of proteins. However, there is a second layer of information present in RNA molecules: parts of the single strand of RNA which carries the genetic code of the virus can fold back onto itself, thereby creating complex 2D and 3D structures. ‘These structures regulate many aspects of the virus life cycle, including its spread and infectivity’, explains Incarnato.
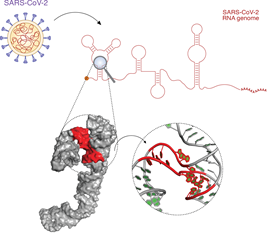
He used a known technique to determine which parts of the RNA are folded. Colleagues from the International Institute of Molecular and Cell Biology (IIMCB) in Warsaw are experts in predicting how the folds organize into a 3D structure. ‘This reveals the presence of pockets, small cavities that could bind small molecules’, says Incarnato. The binding of a small molecule inside such a pocket may hamper the proper functioning of the RNA, and thus affect the virus. ‘We don’t know which pockets are useful for this, but by identifying them, we give drug developers a list of preferential targets which they can investigate.’
The scientists looked especially at structures that are present in different types of coronavirus: these conserved structures are less likely to change through mutations. This is important, as RNA viruses can rapidly mutate to evolve resistance to drugs or vaccines. ‘Some of the 3D structures we looked at are expected not to change much even when there are new mutations in the RNA’, explains Incarnato. This is a clear advantage of drugs designed against the RNA structure.
Other parts of the RNA are not folded. These areas are candidates for so-called antisense oligonucleotide therapy. This means a short artificial DNA molecule is created that will bind to the virus RNA. This double-stranded structure is sensitive to destruction by an enzyme that is naturally present in our cells. Incarnato: ‘Over the last few years, a number of antisense RNA treatments have been approved, so this is a promising field.’
The study was done through a combination of lab work (isolating the RNA and detecting the folds) and computational biology (creating a 3D structure and identifying drug-binding pockets). The results can speed up the search for drugs to combat the pandemic virus. Incarnato and his colleagues are already searching for drug candidates. He is confident that the structures that are described in the paper are sound. ‘We checked our structures against the known structure for different coronaviruses and we found a very good match.’ However, he concludes, the manuscript on the preprint server is work in progress: peer review may alter parts of it. ‘That is how science works, you need your peers to check for details you might have overlooked. And I deeply believe this is essential.’
Reference: Ilaria Manfredonia, Chandran Nithin, Almudena Ponce-Salvatierra, Pritha Ghosh, Tomasz K. Wirecki, Tycho Marinus, Natacha S. Ogando, Eric J Snijder, Martijn J van Hemert, Janusz M. Bujnicki, Danny Incarnato: Genome-wide mapping of therapeutically-relevant SARS-CoV-2 RNA structures. bioRxiv, 15 June 2020
More news
-
10 February 2026
Why only a small number of planets are suitable for life
-
09 February 2026
Can we make the earth spin in the opposite direction?
