Chemical, cellular and pharmaceutical engineering
On this page you may find examples of successful projects developed in the area of chemical, cellular and pharmaceutical engineering by Engineering researchers at the University of Groningen and public and private partners. The list will be constantly updated with new promising initiatives.
Development of novel antimicrobials by bioengineering and synthetic biology
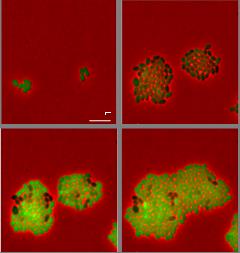
Antibiotic resistance in human pathogens is on the rise and this rise is not met with new approved antibiotics to combat these resistant bacteria. To solve this growing problem of resistance, alternative sources of antibiotics should be explored. The Kuipers group (Molecular Genetics, FSE) investigates several of these sources, particularly the class of ribosomally synthesized, post-translationally modified peptides called lantibiotics. Lantibiotics are potent antimicrobial peptides that undergo extensive posttranslational modifications. In an attempt to improve the spectrum of lantibiotics against Gram-negative microorganisms, we designed different fusions of peptides active against Gram-negative bacteria to the model lantibiotic nisin and parts of nisin that retain the important lipid II binding activitya-c. Thereby, we combined the outer membrane traversing activity of these peptides with the lipid II binding activity of nisin. Several multivalent nisin derivatives were produced by Lactococcus lactis, purified and tested against different bacteria and that some of them showed an increased activity against Escherichia colic . These results present a promising strategy for the rational design and production of novel multivalent lantibiotics targeting Gram-negative pathogens. Moreover, we have used several other approaches to develop novel antimicrobials or to get insight in resistance development; for instance, i) a synthetic biology and bioinformatics approach to efficiently use the large number of sequenced genomes (>4,000) as a resource for novel lantibioticsd-g ii) use of additional heterologously expressed post-translational modification enzymes to hypermodify lantibioticsb,f,h, iii) design and production of novel lantibiotics by ring module- and hinge-variationi, or introduction of non-canonical amino acidsj,k and assessing heteroresistance in human pathogens against lantibiotic derivatives, by using single cell analysesl (Figure).
a Science (2006), bFEMS Microbiol Rev (2016); cFront Cell Dev Biol (2016); dNAR(2013); eBMC Genomics (2016); fACS Synthetic Biology(2016); gNature Chem. Biol. (2015); hACS Synthetic Biology(2013); iFront. Microbiol.(2015); jAmino Acids (2017); kFrontiers Microbiol.(2017)l PNAS(2014);
