Transport through the Nuclear Pore Complex
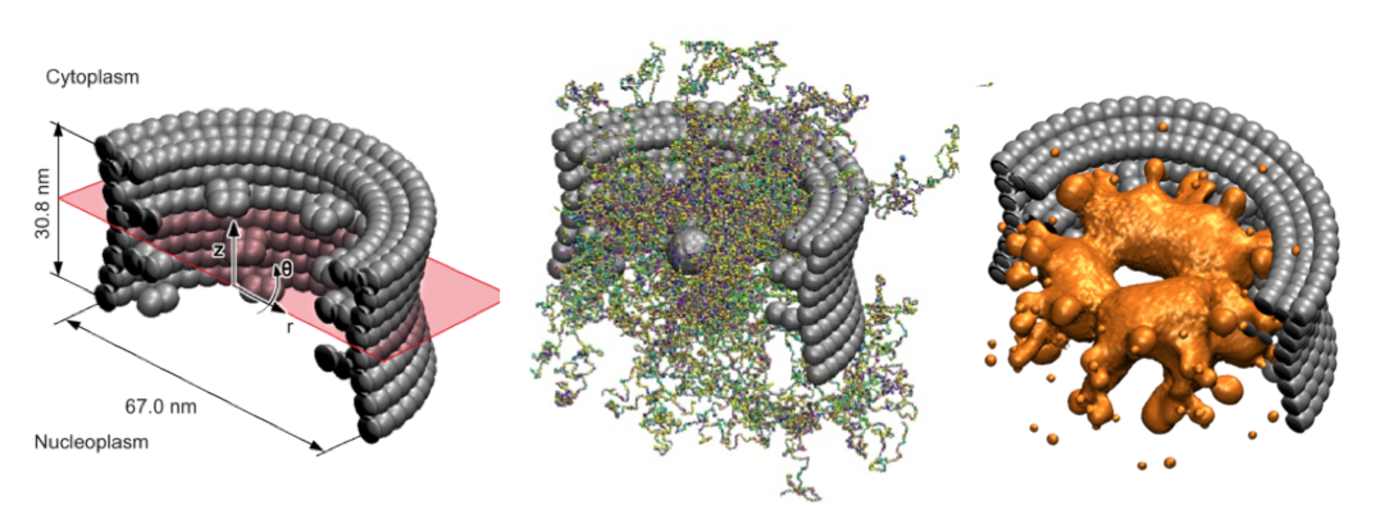
The nuclear pore complex (NPC) is the central gatekeeper that controls the transport of all proteins and RNA molecules into and out of the nucleus of our cells. The NPC is highly selective: Large macromolecules are rejected for transport, unless they are bound to specific transporter molecules (kaps) that travel back and forth between the nucleus and the cytosol. The basic mechanism for NPC transport and selectivity has not yet been resolved, but what is clear, is that intrinsically disordered proteins (FG-Nups) in the core of the NPC play a key role. How these FG-nups actually mediate transport is unclear and subject to intense debate.
To explore this, within the Berendsen Center we have developed an experimentally calibrated one-bead-per-amino-acid molecular dynamics model that enables the molecular modeling of the disordered FG-domain of the NPC and its role in transport. The coarse-grained model distinguishes between all 20 amino acids of the FG-Nups, takes into account hydrophobic and electrostatic interactions between the amino acids, the backbone stiffness of the Nups as well as the screening effect of free ions and polarity of the solvent. The key new feature of the model is that it is fine enough to represent the exact amino-acid sequence of each FG-Nup, but coarse enough to capture the collective behavior of all FG-Nups inside the NPC (containing over 80.000 amino-acids).
We have used the model to resolve the ensemble-averaged protein density distribution in the confined three-dimensional geometry of the NPC. We used umbrella sampling to compute the potential of mean force of transporting particles. We found that the energy barrier for transporting particles increases with particle size and reduces by the presence of hydrophobic binding spots on the particle’s surface, shedding light on the pore’s ability to be a selective gate and a permeability barrier at the same time. Current work is carried out to explore the effect of aging on the structure-function relations of the NPC in collaboration with researchers of ERIBA at the UMCG (Veenhoff Lab).
Key references:
A. Ghavami, E. van der Giessen and P.R. Onck, Coarse-grained potentials for local interactions in unfolded proteins, J. Chem. Theory Comput. 9, 432−440 (2013)
A. Ghavami, L.M. Veenhoff, E. van der Giessen, and P.R. Onck, Probing the disordered domain of the nuclear pore complex through coarse-grained molecular dynamics simulations, Biophys. J. 107, 1393 (2014).
A. Ghavami, E. van der Giessen and P.R. Onck, Energetics of Transport through the Nuclear Pore Complex, PLOS ONE, 11(2), e0148876 (2016).
