The Hansenula polymorpha expression system
Introduction
Hansenula polymorpha (Ogataea polymorpha) is a methylotrophic yeast that can utilize methanol as the sole carbon source. During growth on methanol several peroxisomal enzymes are dramatically induced like alcohol oxidase (AOX), dihydroxyacetone synthase (DHAS) and catalase (CAT). Peroxisomes are highly dynamic organelles that can vary in size and number depending on the carbon source. When grown on glucose, conditions which do not require peroxisomal metabolic pathways, H. polymorpha generally contains a single small peroxisome per cell. However, on methanol H. polymorpha cells proliferate their peroxisomes and these organelles will increase in size (Figure 1). Under certain conditions the total volume fraction of peroxisomes may reach up to 80% of the total cell volume.
The H. polymorpha expression platform
The promoters of the peroxisomal enzymes AOX and DHAS are extremely strong. This property makes H. polymorpha an excellent host for the production of recombinant proteins both for industrial applications as well as for research purposes (e.g. X-ray crystallography). The yeast H. polymorpha is used as a host for the production of several products in the industry. The Biopharmaceutical company Crucell produces a vaccine against hepatitis B (Hepavax-Gene®), interferon alpha 2a for the treatment of hepatitis C and several enzymes that are approved as food or feed additives in this yeast. Another Biotechnological company named ARTES offers R&D contract service for the development of vaccines, bio-pharmaceuticals, bio-similars and enzymes in the yeast H. polymorpha. These examples show that H. polymorpha is an excellent host for the production of heterologous proteins. The main key features why H. polymorpha should be used as a production host are described below.
main key features:
· Highly thermo-tolerant yeast (up to 50°C or more)
· Stable high-copy integration up 120 copies per cell
· Absence of allergens, toxins and viral contaminations
· Large toolbox including inducible strong promoters
· Stable co-expression of multiple genes
· High yields and simple purification methods are available
· Metabolic engineering of whole pathways is possible
Most industrial and laboratory strains are derived from three H. polymorpha strains that were initially isolated from soil, spoiled concentrated orange juice or in the gut of insects. All three strains have been sequenced and two of the sequences are available online.
· Strain DL-1 http://www.ncbi.nlm.nih.gov/genbank/
· Strain NCYC495 leu1.1 http://www.straininfo.net/strains/497657
· Strain CBS 4732 (no sequence available online)
Expression plasmids
In the Department of Molecular Cell Biology a collection of expression vectors have been created in the past 20 years. All plasmids are available for research purpose upon request.
Nomenclature:
All the plasmids are named according to the same nomenclature. In Figure 2 a schematic map is drawn of plasmid pHIPZ4 which is a H. polymorpha integration plasmid containing the strong alcohol oxidase promoter and zeocine as a selection marker.
Example: plasmid pHIPZ4
p = plasmid
HIP = Hansenula Integration Plasmid
Z= Zeocin (marker used for integration in yeast)
4 = Promoter used in this example AOX
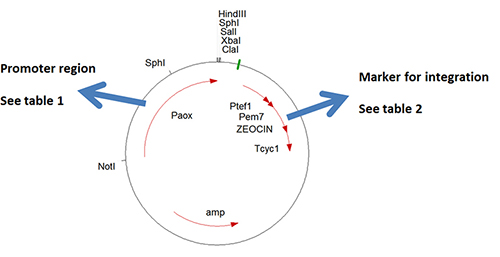
Table 1. Available integration plasmids
|
Vector number |
promoter |
Available vectors |
|
1 |
AOX |
pHIPX1 (not used for routine cloning) |
|
2 |
AOX |
pHIPX2 (not used for routine cloning) |
|
3 |
AOX |
pHIPX3 (not used for routine cloning) |
|
4 |
AOX |
pHIPX4, pHIPZ4, pHIPN4, pHIPB4, pHIPH4, pHIPA4, pHIPM4, pHIPK4 |
|
5 |
AMO |
pHIPX5, pHIPZ5, pHIPN5, |
|
6 |
PEX3 |
pHIPX6, pHIPZ6 |
|
7 |
TEF |
pHIPX7, pHIPZ7, |
|
8 |
TEF2 |
pHIPX8 |
|
9 |
CAT |
pHIPX9 |
|
10 |
PEX14 |
pHIPX10 |
|
11 |
PEX4 |
pHIPX11 |
|
12 |
PEX5 |
pHIPX12 |
|
13 |
PEX19 |
pHIPX13 |
|
14 |
INP2 |
pHIPX14 |
|
15 |
DHAS |
pHIPZ15 |
|
16 |
LYS5 |
pHIPA16-SFP (no empty plasmid) |
|
17 |
PEX11 |
pHIPZ17-Nia |
|
18 |
ADH1 |
pHIPZ18 eGFP-SKL |
Description of the most important promoters.
PAOX very strong promoter that is repressed on glucose and induced during growth on methanol
PTEF constitutive promoter, same expression on glucose and methanol
PAMO promoter induced by primary amines (methylamine, ethylamine) and fully repressed by ammonium sulphate
PADH1 constitutive promoter, comparable to TEF on glucose but higher on methanol
Available markers
For genetic engineering of H. polymorpha strains both auxotrophic markers as well as dominant markers can be used (Table 2).
Table 2 Markers for integration in H. polymorpha
|
Marker |
Description |
|
Sc LEU2* |
Leucine auxotrophy |
|
Hp URA3 |
Uracil auxotrophy |
|
Hp ADE11 |
Adenine auxotrophy |
|
Hp MET6 |
Methionine auxotrophy |
|
Sh-ble |
Zeocin resistance |
|
Sn-nat1 |
Nourseothricin resistance |
|
Kp-hph |
Hygromycin B resistance |
|
Tn-KanMX |
G418/Geneticin resistance |
|
Sv-Pat |
Bialaphos resistance |
*Can be used for multi-copy integration
Strain for efficient integration
Non-Homologous end joining recombination (NHEJ) is important for double strand break repair, but also causes a high frequency of random integration. The Ku heterodimer binds to DNA and is required for NHEJ. Ku consists of two subunits, called Ku70 and Ku80.
We created a H. polymorpha strain lacking the Ku80 subunit (a YKU80 knockout strain). When this strain is used for directed integration, the number of transformants decreases, but the percentage of correct integration dramatically increases (Saraya et al., FEMS Yeast Res 12 (2012) 271–278). Therefore this strain is generally used to create knockout strains.
Strain: NCYC495 yku80 leu1.1
Strain request
All strains and plasmids are available for research purposes upon request.
Contact person: A.M. Krikken
Nijenborgh 7
9747 AG Groningen
050 – 363 2177
a.m.krikken rug.nl
Links
· CRUCELL
· PHARMEDARTIS
