Peroxisomal protein export
VIDI fellowship awarded to Dr. Chris Williams
Due to their vital role in cellular metabolism, peroxisomes need to adapt their protein content to changes in metabolic needs. For this reason, the ability to selectively export proteins, for degradation or targeting to other cellular compartments, can be invaluable. In contrast to other cell organelles, only a few examples of peroxisomal protein export are known and the details behind how and why proteins are exported out of peroxisomes remain largely unexplored ( Williams 2014 ).
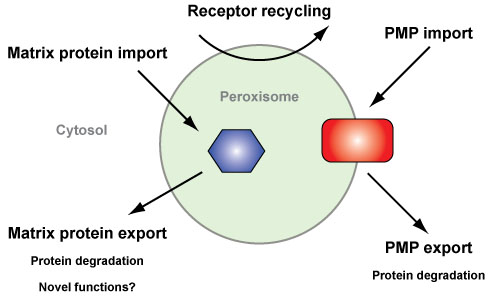
Research in the export group (Dr. Chris Williams, Chen Xin, Natasha Danda and Srishti Devarajan) focusses on the ubiquitin-dependent export and degradation of peroxisomal membrane proteins (PMPs). Our recent work demonstrates that the PMP Pex3p is specifically exported from peroxisomes and degraded upon changes to growth conditions, a process that is regulated by ubiquitination of Pex3p ( Williams and van der Klei 2013 ). Ubiquitination (Figure 2), the attachment of the protein ubiquitin to a substrate, often targets proteins for proteasome-mediated ( Tanaka 2013 ). Furthermore, Pex3p ubiquitination requires Pex2p and Pex10p; two ubiquitin ligases already implicated in peroxisomal ubiquitination events ( Williams et al, 2008 ; Platta et al, 2009 ).
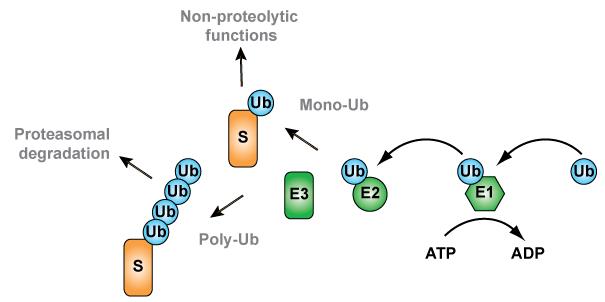
Our research focusses on the following:
- By employing yeast genetics, together with various biochemical approaches, we aim to determine additional proteins involved in Pex3p ubiquitination.
- Identify other substrates of the PMP export pathway, using a combination of proteomics and state-of-the-art fluorescence microscopy techniques.
- Investigating the impact of PMP export on cell function, through the use of physiological approaches, combined with advanced electron microscopy/tomography and confocal laser scanning microscopy.
- With purified proteins, we follow ubiquitination reactions in vitro using biochemical and more recently, single molecule techniques, in order to investigate the molecular mechanisms underlying PMP ubiquitination.
| Last modified: | 19 January 2016 1.43 p.m. |
