Evolutionary systems biology
| Research | People | Publications |
TRÊS is currently strengthening its research activities at the interface of evolutionary biology and the molecular life sciences. As a start, we will investigate the evolution of gene- and protein-interaction networks. Understanding how such networks function is prerequisite to unlocking the wealth of molecular data that has become available.
The work in our group concentrates on bacteria, which are currently most amenable to an evolutionary systems biology approach. Using bacterial chemotaxis, natural competence, biofilm formation and metabolic switching between carbon sources as model systems, we ask how the complexity of biomolecular interactions influences the dynamics of evolutionary adaptation, and how it constrains or facilitates phenotypic diversification through either plasticity, bet-hedging or the establishment of genetic polymorphism. To answer these questions, we rely on theoretical modeling techniques from systems biology and traditional evolutionary theory, as well as on evolution experiments (in collaboration with the Molecular Genetics group).
A second research objective is to utilize evolutionary insights to develop new model-reduction strategies for systems biology. Preliminary results suggests that complex interaction networks can often be reduced to functionally equivalent simpler networks that are reachable by a series of nearly neutral evolutionary steps each involving only a local rewiring of the network (Figure 1). This observation is consistent with the idea that evolution typically first converges on a simple solution for a problem, which is subsequently refined by a process of tinkering.
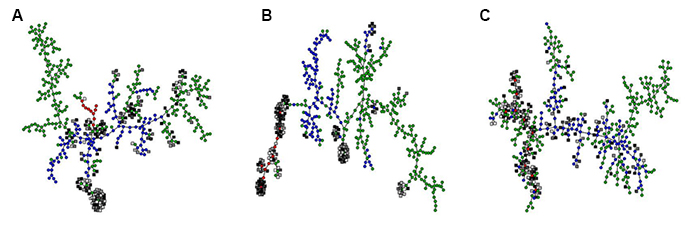
In a model for the evolution of phenotypic plasticity, we studied how a signal-transduction network of interacting proteins could process information from the environment in order to realize an optimal pattern of phenotypic adjustment over the lifetime of the organism. This is a complex optimization task with many (locally) optimal solutions, each of which is represented by a node in A-C. Some of the optimal solutions can be implemented by a simple network of three interacting proteins (red circles), others require four (blue circles) or up to five proteins (green circles). We also generated solutions by the trial-and-error process of evolution (grey squares). These were clustered together with the optimal solutions based on molecular similarity (i.e., similar network structure; A), phenotypic similarity (i.e., similar developmental trajectories of the phenotype; B) and evolutionary similarity (i.e., reachability by a series of nearly neutral small-scale network modifications; C). The resulting graphs show that the evolved solutions are nearly-neutral equivalents of the simplest possible network architectures capable of implementing age-dependent plasticity (C). This equivalence is also apparent at the phenotypic level (B), but not at the level of network structure (A), because neutral and redundant interactions obscure which interactions in the network are important for its function. Both (A) and (C) recover the same clusters of structurally similar optimal solutions, but only evolutionary similarity (C) predicts which ones of these can be reached by evolution.
