Peroxisomal membrane protein degradation
Peroxisomes are organelles that play a critical role in cellular metabolism, providing compartments where metabolic pathways are contained and controlled. Since peroxisomes do not contain DNA, they post-translationally import all the matrix and membrane proteins required for function, demonstrating the vital role protein transport plays in peroxisome function. The importance of peroxisomes for cell vitality is underlined by the developmental brain disorders caused by peroxisome malfunction. Disturbances in peroxisome function also contribute to ageing.
In contrast to other organelles, little is known about how PMPs are removed from the peroxisomal membrane and targeted for degradation . PMP degradation may serve a quality control function for the degradation of damaged or misfolded PMPs but it may also allow the removal of redundant PMPs, to regulate peroxisome function. Research in our group (Chris Williams; Srishti Devarajan; Xin Chen) focusses on investigating the mechanisms and functions of PMP degradation.
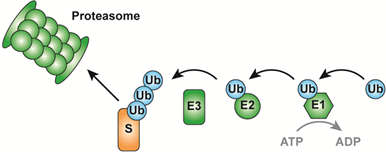
Protein degradation via the ubiquitin-proteasome system
The ubiquitin-proteasome system (UPS, Figure 1) is the major protein degradation pathway in eukaryotic cells. Attachment of the 8 kDa protein ubiquitin (Ub) to a substrate protein (S) is an ATP dependent process, requiring three distinct enzymes. First, the ubiquitin activating enzyme (E1) activates ubiquitin. Next, the activated ubiquitin is transferred to the active site cysteine residue of an ubiquitin conjugating enzyme (E2). Finally, an ubiquitin ligase (E3) allows conjugation of ubiquitin to the substrate. The ubiquitin attached to the substrate can itself become a substrate for ubiquitination, resulting in the formation of ubiquitin chains. Such Ub chains can target substrates for degradation via the proteasome.
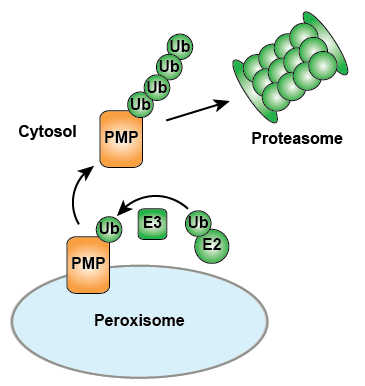
Investigating UPS mediated degradation of PMPs
We recently demonstrated that PMPs can be selectively degraded via the UPS and identified E2 and E3 enzymes that play a role in PMP ubiquitination and degradation. These data demonstrate that a UPS dependent pathway can selectively target PMPs for cytosolic degradation (Figure 2). Additionally, we have gained detailed insights into the molecular mechanisms of PMP degradation and collaborate on developing techniques to follow ubiquitination at the single molecule level (Wloka et al 2017).
By combining state-of-the-art microscopy and biochemical techniques with structural biology and in vitro approaches, we aim to;
- provide further insights into the degradation of the PMPs Pex3p and Pex13p
- identify additional substrates of the PMP degradation pathway
- investigate the impact of PMP export on peroxisome function and cell health
- uncover further the mechanisms that control PMP ubiquitination and degradation
Former group members
Natasha Danda – lab technician (2015-2017). Currently Research Engineer at the Institut du Cerveau et de la Moelle épinière (ICM) in Paris, France.
Former undergraduate students
Inge Bles (2017)
Alwin Slager (2017)
Femke Heimstra (2016)
Nieck van der Heide (2016)
Joana Neto-Gomes (2015)
Alfred Zwikstra (2015)
Dewi van Gelder (2015)
Hans Wolsink (2014)
