
Genetics of complex immune-related diseases
Prof. Cisca Wijmenga’s group performs research in the field of complex genetics, focusing on immune-related diseases. In complex diseases, a large number of genetic factors, acting together with non-genetic (environmental) factors, are responsible for the development of the disease. Important examples include irritable bowel disorders (IBD, comprising Crohn’s disease and ulcerative colitis (UC)), coeliac disease (CD, gluten intolerance), asthma, diabetes, multiple sclerosis and rheumatoid arthritis (RA). The work on gastrointestinal disorders has led to functional genomics studies into the gut microbiome. Further work is also being done on the functional genomics of fungal and other infections.
This research is mostly carried out using genome-wide, hypothesis-free approaches. We have a large and unique collection of patient materials, many collaborative partners, and make use of cutting-edge technology and big data analyses. This means the group is in an excellent position to translate genetic knowledge to the underlying molecular mechanisms of disease. See also Systems genetics and Genomic Coordination Center
See a keynote presentation on the microbiome work, Our second genome in health and disease, by Cisca Wijmenga given to 27th European Congress of Clinical Microbiology and Infectious Diseases (ECCMID 2017, Vienna, Austria) on 24 April 2017 and a short interview with her.
Recent papers:
The MHC locus and genetic susceptibility to autoimmune and infectious diseases Matzeraki et al. Genome Biology 2017
The influence of proton pump inhibitors and other commonly used medication on the gut microbiota Imhann et al. Gut Microbes 2017
Disease variants alter transcription factor levels and methylation of their binding sites Bonder MJ et al. Nat Genet 2017
Identification of context-dependent expression quantitative trait loci in whole blood Zhernakova DV et al. Nat Genet 2017
Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity Schirmer M et al. Cell 2016
Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease Imhann et al. Gut 2016
Inter-individual variability and genetic influences on cytokine responses to bacteria and fungi Yang Li et al. Nature Medicine 2016
Proton pump inhibitors affect the gut microbiome Imhann F et al. Gut 2016;65 (5), 740-748
Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity A Zhernakova et al. Science 2016;352 (6285), 565-569
Population-level analysis of gut microbiome variation Falony G et al. Science 2016;352 (6285), 560-564
Refined mapping of autoimmune disease associated genetic variants with gene expression suggests an important role for non-coding RNAs Ricaño-Ponce I et al. J Autoimmun. 2016 Apr;68:62-74
Functional implications of disease-specific variants in loci jointly associated with coeliac disease and rheumatoid arthritis Gutierrez-Achury J et al. Hum Mol Genet. 2016 Jan 1;25(1):180-90
The genome revolution and its role in understanding complex diseases Hofker MH, Fu J, Wijmenga C. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease, 2014
Mapping of immune-mediated disease genes Ricaño-Ponce I, Wijmenga C. Ann rev genomics and hum genetics 2013;14, 325-353
(1) Coeliac disease
Coeliac disease is an intolerance for dietary gluten, a protein found in one of our main foods: grains. The disease is characterized by an immune-mediated damage to the small intestinal mucosa (“flat mucosa”). It is the most frequently occurring food intolerance, affecting some 1% of the population. At the same time, the disease often goes unrecognized by doctors, because of the broad range of systemic and sometimes vague complaints – varying from osteoporosis to infertility to motor and balance disorders (ataxias) to general fatigue; a correct diagnosis is made in fewer than 10% of cases. In addition these patients often suffer from other autoimmune diseases like type 1 diabetes or thyroid problems.
See also 'The genetics of coeliac disease', Courtesy of International Innovation – a leading scientific dissemination service and the Dutch Celiac Disease Consortium's (CDC) brochure (in Dutch)

Thanks to the many genetic breakthroughs made in the past few years, coeliac disease is now one of the best understood complex diseases and it is showing the way for other diseases. This research has provided a revelation in that the disease genes identified for coeliac disease are not unique to that disease, but often shared with other immune-related diseases, such as Crohn’s disease, rheumatoid arthritis and type 1 diabetes.
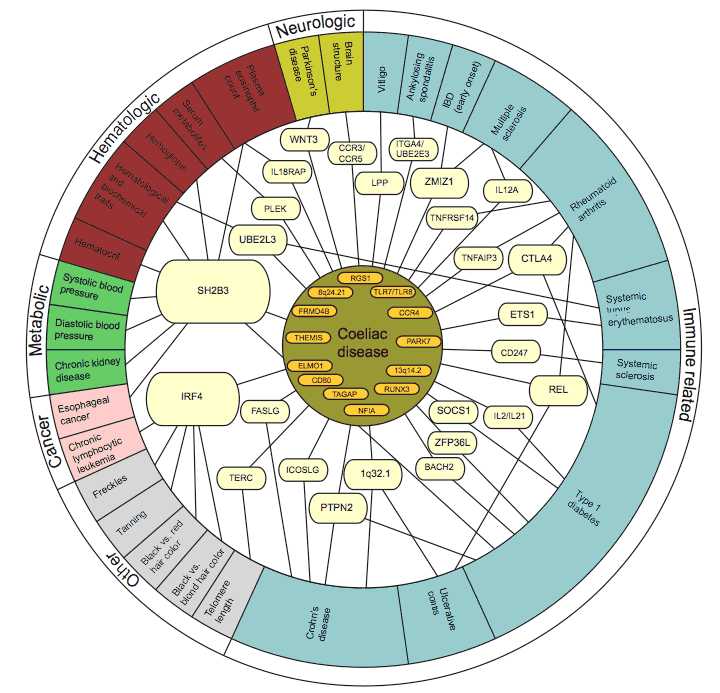
Importantly, the research has already yielded many new pointers to the mechanisms underlying the development of coeliac disease and the overlapping immune-related diseases.
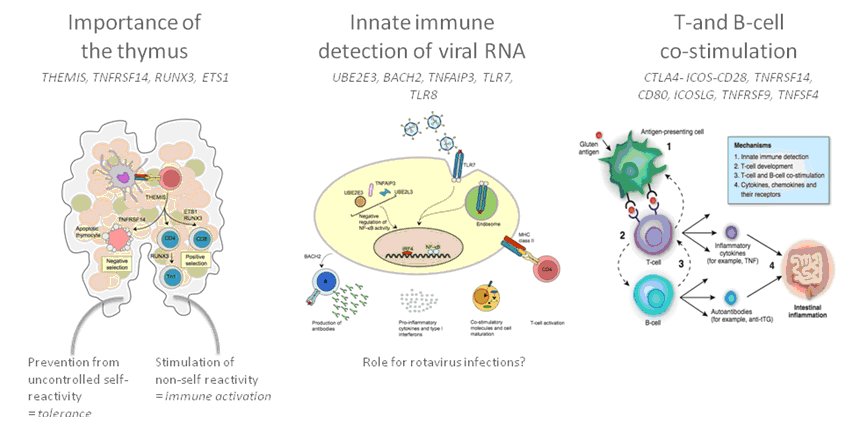
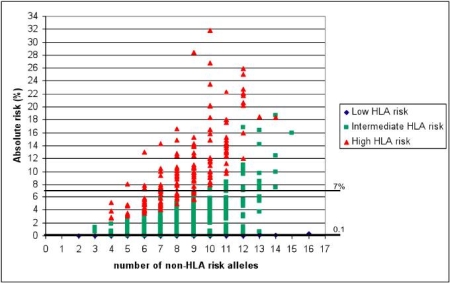
The genes found by Wijmenga’s group can explain approximately 40% of the genetics of coeliac disease. Now we want to find a way to use this genetic information to trace potential patients. This target has come a step closer with recent results, as shown by Romanos et al. (2010), in which we outline how specific DNA tests can be used in a model to identify people with a higher risk of developing coeliac disease.
The central question that we want to answer in the coming years is ‘how does variation in a large number of different genetic factors together lead to disease?’ And then: ‘how can we use this knowledge to prevent, better diagnose or treat diseases?’ To explore these questions we will make use of gene expression data in different cells and tissues from coeliac disease patients and controls. This data will also be integrated with genetic and phenotypical data (and, in due course, with environmental factors).
TIFN prize
The Joop Roels Impact Award 2013 was presented to Prof. Cisca Wijmenga and her team for their research project on the ‘Validation of biomarkers’ at the Top Institute Food and Nutrition’s 2014 annual conference, held in Utrecht on 22 May 2014. TIFN’s prizes aim to recognise the best scientists and scientific achievements and to highlight the societal and industrial impact of the research carried out. This award consists of the sum of €1,500 and a work of art in glass.
Selected references
The influence of a short-term gluten-free diet on the human gut microbiome. Bonder MJ, et al. Genome Med. 2016;8(1):45.
Understanding Celiac Disease by Genomics. Withoff S, Li Y, Jonkers I, Wijmenga C. Trends Genet. 2016;32(5):295-308.
A large variety of clinical features and concomitant disorders in celiac disease - A cohort study in the Netherlands. Spijkerman M, et al. Dig Liver Dis. 2016;48(5):499-505.
Contrasting the Genetic Background of Type 1 Diabetes and Celiac Disease Autoimmunity Gutierrez-Achury J, et al. Diabetes Care. 2015;38 Suppl 2:S37-44
Genetics of celiac disease Ricaño-Ponce I et al. Best Practice & Research Clinical Gastroenterology 2015,
Abstract: New insights into the underlying molecular pathophysiology of celiac disease (CeD) over the last few years have been guided by major advances in the fields of genetics and genomics. The development and use of the Immunochip genotyping platform paved the way for the discovery of 39 non-HLA loci associated to CeD, and for follow-up functional genomics studies that pinpointed new disease genes, biological pathways and regulatory elements. By combining information from genetics with gene expression data, it has become clear that CeD is a disease with a dysregulated immune response, which can probably occur in a variety of immune cells. This type of information is crucial for our understanding of the disease and for providing leads for developing alternative therapies to the current gluten-free diet. In this review, we place these genetic findings in a wider context and suggest how they can assist the clinical care of CeD patients.
Deelen P et al. Calling genotypes from public RNA-sequencing data enables identification of genetic variants that affect gene-expression levels. Genome Medicine 2015, pdf
RNA-sequencing (RNA-seq) is a powerful technique for the identification of genetic variants that affect gene-expression levels, either through expression quantitative trait locus (eQTL) mapping or through allele-specific expression (ASE) analysis. [...] By deriving and imputing genotypes from RNA-seq data in the public domain, it is possible to identify both eQTLs and ASE effects. Given the exponential growth of the number of publicly available RNA-seq samples, we expect this approach will become especially relevant for studying the effects of tissue-specific and rare pathogenic genetic variants to aid clinical interpretation of exome and genome sequencing.
Gutierrez-Achury J et al.
Fine mapping in the MHC region accounts for 18% additional genetic risk for celiac disease. Nat Genet. 2015;47(6):577-8.
Abstract: Although dietary gluten is the trigger for celiac disease, risk is strongly influenced by genetic variation in the major histocompatibility complex (MHC) region. We fine mapped the MHC association signal to identify additional risk factors independent of the HLA-DQA1 and HLA-DQB1 alleles and observed five new associations that account for 18% of the genetic risk. Taking these new loci together with the 57 known non-MHC loci, genetic variation can now explain up to 48% of celiac disease heritability. Press release
Wijmenga C, Gutierrez-Achury J, Celiac Disease Genetics: Past, Present and Future Challenges, Journal of pediatric gastroenterology and nutrition 2014; 59, S4-S7.
Abstract: In the past few years there has been enormous progress in unraveling the genetic basis of celiac disease (CD). Apart from the well-known association to HLA, there are currently 40 genomic loci associated to CD. Most of these loci show pleiotropic effects across many autoimmune diseases and highlight the importance of a dysregulated immune system in the predisposition to CD. It is still too early, however, to use genetics in clinical practice for predicting individual risk. The major challenge for the future is to translate genetic findings into a better understanding of the underlying disease mechanism and to design new ways to treat CD and prevent its development.
Cénit MC et al. Rapidly expanding knowledge on the role of the gut microbiome in health and disease. Biochim Biophys Acta. 2014;1842(10):1981-1992
Many studies have indicated that the genomes of our gut microbiota, known as the gut microbiome or our "other genome", could play an important role in immune-related, complex diseases, and growing evidence supports a causal role for gut microbiota in regulating predisposition to diseases. A comprehensive analysis of the human gut microbiome is thus important to unravel the exact mechanisms by which the gut microbiota are involved in health and disease. Recent advances in next-generation sequencing technology, along with the development of metagenomics and bioinformatics tools, have provided opportunities to characterize the microbial communities. Furthermore, studies using germ-free animals have shed light on how the gut microbiota are involved in autoimmunity. In this review we describe the different approaches used to characterize the human microbiome, review current knowledge about the gut microbiome, and discuss the role of gut microbiota in immune homeostasis and autoimmunity.
Hofker MH et al. The genome revolution and its role in understanding complex diseases. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease 2014; 1842 (10), 1889-1895.
Zhernakova A et al. Clinical implications of shared genetics and pathogenesis in autoimmune diseases. Nat Rev Endocrinol. 2013;9(11):646-59. Review.
Zhernakova et al. Detecting shared pathogenesis from the shared genetics of immune-related diseases. Nat Rev Genet. 2009; 10(1):43-55. Review.
(2) IBD
Inflammatory bowel disease (IBD) comprises Crohn’s disease and ulcerative colitis, two of the most common forms of chronic inflammation of the intestine. IBD affects approximately one in 250 patients in the Western world. The disease onset is mostly in the second or third decade of life and because of symptoms like bloody diarrhoea, abdominal pain and anaemia it has a high impact on the patient’s quality of life. IBD is a multifactorial disease in which an aberrant immune response occurs to commensal microflora in a genetically susceptible host.
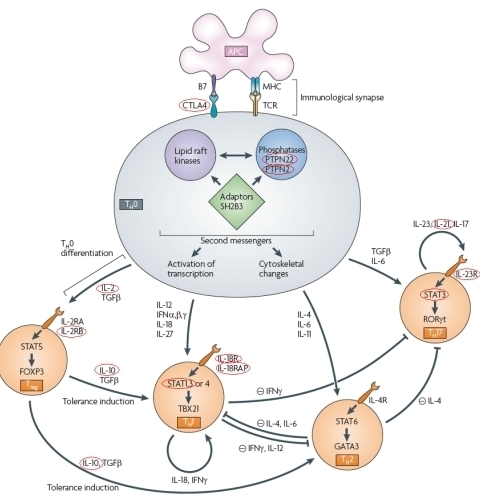
10 years after the first associated locus was found for Crohn’s disease (Hugot et al. 2001), intensive research has identified 30 loci associated to Crohn’s disease (Barrett et al. 2010), and 18 loci associated to ulcerative colitis (McGovern et al. 2010). However, these still only explain approximately 10% of the heritability. Many of the associated loci are shared between both types of IBD or with other immune-related diseases (Festen et al. 2009; Zhernakova et al. 2009), while some are disease-specific. Most of the genes are involved in T-cell signalling (see figure). New approaches are being developed to solve the puzzle of the remaining heritability (Fransen et al. 2010). To discover further pathways involved in IBD, more associated genes need to be identified and their role in the pathogenesis needs to be determined.
References
A novel biomarker panel for irritable bowel syndrome and the application in the general population. Mujagic Z, et al. Sci Rep. 2016;6:26420.
High-density mapping of the MHC identifies a shared role for HLA-DRB1*01:03 in inflammatory bowel diseases and heterozygous advantage in ulcerative colitis. Goyette et al. Nat Genet. 2015;47(2):172-9. doi: 10.1038/ng.3176.
Fransen K et al. Correlation of genetic risk and messenger RNA expression in a Th17/IL23 pathway analysis in inflammatory bowel disease. Inflamm Bowel Dis. 2014;20(5):777-82.
Fransen K, Inflammatory bowel disease: the genetic background and beyond, PhD thesis, University of Groningen, 2014, full text available
Liu JZ et al. Dense genotyping of immune-related disease regions identifies nine new risk loci for primary sclerosing cholangitis. Nat Genet. 2013;45(6):670-5. doi: 10.1038/ng.2616.
Jostins L et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease, Nature 2012;491 (7422), 119-124
Zhernakova A, et al. Detecting shared pathogenesis from the shared genetics of immune-related diseases. Nat Rev Genet. 2009;10(1):43-55. Review.
(3) COPD
Chronic obstructive pulmonary disease (COPD) is currently the third leading cause of death worldwide, according to the WHO, and is expected to become an even more serious problem in upcoming decades. COPD is a late-onset disease mainly caused by tobacco smoking, but air pollution and other environmental factors can also contribute to its development. The lung tissue of COPD patients becomes inflamed and destroyed, which leads to serious breathing problems and may lead to death.
Studies have proved there is a genetic component involved in the development of COPD, but more than two decades of using candidate gene approaches has resulted in many spurious results. A meta-analysis of these studies proved only two out of 12 loci to be associated with COPD (Smolonska et al., 2009). When the first genome-wide association (GWA) study on COPD was published (Pillai et al., 2009), none of the candidate genes were among the most associated genes found (nor in the following study (Cho et al., 2010)). The GWA studies have pointed to new genes (CHRNA3/5, HHIP, FAM13A), but their role in COPD is unknown and the mechanisms for their action need to be determined by functional studies.
References
Dijkstra AE et al. Novel genes for airway wall thickness identified with combined genome-wide association and expression analyses. Am J Respir Crit Care Med. 2015;191(5):547-56.
Dijkstra AE et al. Dissecting the genetics of chronic mucus hypersecretion in smokers with and without COPD. Eur Respir J. 2015;45(1):60-75.
Smolonska J, Koppelman GH, Wijmenga C, et al. Common genes underlying asthma and COPD? Genome-wide analysis on the Dutch hypothesis, European Respiratory Journal, 2014;44(4):860-72.
Smolonska, J. “The many faces of COPD”. PhD thesis, University of Groningen, 2013 (full text via link)
| Last modified: | 07 February 2020 3.01 p.m. |
