Research Highlight
Prof. dr. Siewert Jan Marrink
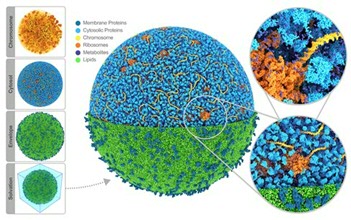
[1] Stevens J.A., Grünewald F., van Tilburg P.A.M., König M., Gilbert B.R., Brier T.A., Thornburg Z.R., Luthey-Schulten Z. & Marrink S.J. (2023). Molecular dynamics simulation of an entire cell. Frontiers in Chemistry 11: doi:10.3389/fchem.2023.1106495Cellular membranes contain hundreds of different lipid types and are crowded with membrane proteins. To understand the protein-lipid interplay in such a complex environment, a high-throughput, multi-scale modeling pipeline has been established by the Marrink group, which is based on the locally developed MARTINI force field. This allows modeling of the interaction of large ensembles of lipids and proteins, giving rise to emergent behavior underlying basic cellular processes such as fusion, fission, transport, and signaling.
A recent breakthrough in this area is the realistic model of an entire cell [1], the JCVI-Syn3a minimal cell (see illustration). Here, for the first time, the full complexity of an entire cell is captured in near-atomic detail. The model contains 60,887 soluble proteins (light blue), 2,200 membrane proteins (blue), 503 ribosomes (orange), a single 500 kbp circular dsDNA (yellow), 1.3 million lipids (green), 1.7 million metabolites (dark blue), 14 million ions (not shown) and 447 million water beads (not shown) for a total of 561 million beads, representing more than six billion atoms - a current world record for a biomolecular system, both in size and complexity.
| Last modified: | 26 October 2023 2.17 p.m. |
