Highlight
Dr. Danny Incarnato
Structural and functional dynamics of RNA
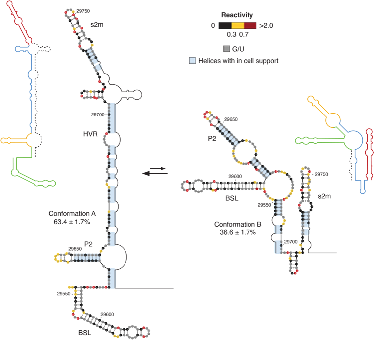
RNA structures are highly diverse and dynamic. Inside the cell, the same RNA is present in multiple identical copies, but not all of them will fold into the same structure. Rather, many alternative structures (or conformations) of the same RNA coexist in a dynamic equilibrium. In addition to that, each conformation is not static over time, but it can interconvert between any other potential conformation for that RNA. This structurally heterogeneous set of RNAs is commonly referred to as an ensemble.
Studying RNA structure ensembles in living cells represents a non-trivial challenge. A recent breakthrough in this area has been the development of the DRACO (Deconvolution of RNA Alternative COnformations) method[1].
The method allows deconvolving the different conformations making up the ensemble, starting from chemical probing data. Its application to the RNA genome of the SARS-CoV-2 revealed the existence of substantial structural heterogeneity at the level of key regulatory regions of the viral genome (see illustration).
By combining DRACO and other cutting-edge methods, the group led by Danny Incarnato aims at providing the first complete map of the RNA structurome (the full set of structures populated by the transcriptome) of a living cell.
[1] Morandi E, Manfredonia I, Simon LM, Anselmi F, van Hemert MJ, Oliviero S & Incarnato D. (2021) Genome-scale deconvolution of RNA structure ensembles. Nature Methods 18: 249-252 DOI: https://doi.org/10.1038/s41592-021-01075-w
| Last modified: | 25 September 2023 1.07 p.m. |
