Research Facilities
Equipment
The following equipment and facilities are available in the Membrane Enzymology group:
- Well-equipped laboratories for recombinant DNA and protein chemistry work
- Well-equipped laboratories for radiochemical work
- Computer-controlled bioreactors for cell cultivation at 1-2 L and 10-12 L scales
- In situ autoclavable 60 L Bioreactor
- High speed centrifuges for volumes up to 2 x 6 L (Beckman Avanti high performance);high speed centrifuges for smaller volumes; Ultracentrifuges (Optima LE-80k, Optima MAX-E and TLX-100); shared with the Biotechnology group
- Optima XL-I Analytical ultracentrifuge with UV/Vis and Interference Optics
- Two Äkta protein purification platforms
- Wyatt Optilab multi-angle light-scattering equipment (coupled to size-exclusion chromatography platform)
- EttanTM DIGE two-dimensional gel electrophoresis equipment, incl. Imaging system (Typhoon 9400) and EttanTM spot picker
- Nanoflow multidimensional liquid chromatography platform (Ettan MDLC) coupled to nanoflow spotter (Probot)
- MALDI-TOF/TOF mass spectrometers (Applied Biosystems 4700/4800 proteomics analyzer)
- LTQ-Orbitrap XL (Thermo Scientific)
- Carry 100 UV/Vis Spectrophotometer
- Applied Photophysics Stopped-flow Spectrophotometer; shared with the Biotechnology group
- SLM Aminco SPF500 Spectrofluorimeter
- SPEX Fluorolog 322 Fluorescence Spectrophotometer
- Ultrafast Polychromatic Fluorescence Lifetime Measuring System equipped with Streak Camera and Time-correlated Single Photon Counting Detector; shared with the Laser Spectroscopy group
- AVIV Cicular Dichroism spectrophotometer
- Microcal MCS Differential Scanning Calorimeter (DSC) and Isothermal Titration Calorimeter (ITC)
- Patch clamp set-up for recording of single channelion conductances; shared with BioMaDe Technology Foundation
- Dual-color laser scanning confocal microscope with three laser beams (488 nm, argon ion laser, Spectra-Physics, and 530 and 633 nm, He-Ne lasers, JDS Uniphase); shared with the Laser Spectroscopy group.
Mass spectometry facility
Two MALDI-TOF/TOF instruments (4700 and 4800 Proteomics Analyzer) and a linear ion trap LTQ-Orbitrap XL (Thermo Scientifics) are available at the Membrane Enzymology group. These instruments are primarily used for projects running in our group and for collaborators within the Netherlands Proteomics Centre.
Measurements for outside groups can be performed after assessment of suitability of the proposed experiment (contact Fabrizia Fusetti or Bert Poolman, see below). The projects of primary interest are membrane protein and proteomics-related research, in particular where MS/MS measurements are required for identification or quantitation of protein samples. For regular MALDI-TOF applications, where no MS/MS is required, please refer to the MS Core Facility (Hjalmar Permentier).
Services and projects undertaken will be on a collaboration basis and are expected to result in co-authorship. Charges include consumables costs only. For academic groups, an hourly rate of € 30 is charged for instrument use (both LC and MS). A time estimate can be provided after discussion of the experimental details.
For more information please refer to the guidelines of the Netherlands Proteomics Centre
Contact details:
Fabrizia Fusetti (phone 050.3634597)
Hjalmar Permentier
Bert Poolman
Optical Microscopy
- Cellular imaging: we use a dual-color confocal microscope for time-lapse imaging of dynamic processes in cells or GUVs.
- Protein & lipid diffusion: the diffusion of lipids1,2 and proteins 2,3 is measured by fluorescence correlation spectroscopy (FCS) and fluorescence-recovery after photobleached (FRAP)-based methods.
- Transport through membrane pores: a method termed dual-color fluorescence burst analysis (DCFBA) has bee developed to probe (macro)molecular transport through mechanosensitive channels and peptide pores. Selective-FRAP is used to probe the in situ transport through the yeast nuclear pore complex.
- Kahya, N., Scherfeld, D., Bacia, K., Poolman, B., and Schwille, P. (2003)
Probing lipid mobility of raft-exhibiting model membranes by Fluorescence Correlation Spectroscopy.
J. Biol. Chem., 278, 28109-28115. - Doeven, M.K., Folgering, J.H.A., Krasnikov, V., Geertsma, E.R., van den Bogaart, G., and Poolman, B. (2005) Distribution, Lateral Mobility and Function of Membrane Proteins Incorporated into Giant Unilamellar Vesicles. Biophys. J., 88, 1134-1142.
- van den Bogaart, G., Hermans, N., Krasnikov, V., and Poolman, B. (2007) Protein mobility and diffusive barriers in Escherichia coli: consequences of osmotic stress. Molec. Microbiol., 64, 858-871 .
Enzymology & Protein Chemistry
- Protein purification: the purification of membrane proteins is routinely done by metal-affinity chromatography, followed by size-exclusion chromatography.1,2
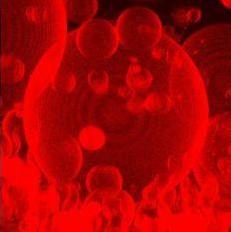
- Membrane reconstitution: protocols have been developed to incorporate integral and peripheral membrane proteins into large unilamellar vesicles (LUVs) and giant-unilamellar vesicles (GUVs, see picture).3
- Ligand binding and substrate translocation: assays have been developed to probe ligand binding and translocation reactions in situ and in vitro.1,2,3,4
-
Poolman, B., Doeven, M.K., Geertsma, E.R., Biemans-Oldehinkel, E., Konings, W.N., and Rees, D.C. (2005) Functional analysis of detergent-solubilized and membrane-reconstituted ABC Transporters. Meth. Enzymol., 400, 429-459.
-
Membrane protein purifation protocol (link to pdf)
-
Geertsma, E.R., Mahmood, N.A.B., Schuurman-Wolters, G.K., and Poolman, B. (2007) Membrane reconstitution of ABC transporters and assays of translocator function. Nature Protoc., in press.
-
Duurkens, R.H., Tol, M.B., Geertsma, E.R., Permentier, H.P., and Slotboom, D.J. (2007) Flavin binding to the high affinity riboflavin transporter RibU. J. Biol. Chem., 282, 10380-10386.
| Last modified: | 29 May 2019 3.02 p.m. |
