Location is what counts for bacterial genes
Bacteria carry their genes on a circular chromosome. Because of the way this chromosome is copied before cell division, the genes’ chromosomal location can affect their expression. University of Groningen microbiologists Jan-Willem Veening and Jelle Slager describe how gene location influences bacterial processes in a review that was published in Trends in Microbiology on 27 Juni (corrected proofs).
They say that important discoveries are made not with shouts of ‘Eureka!’ but rather with a mumbled ‘that’s odd’. In a 2014 paper , the Veening group described that treating a streptococcus bacterium with certain antibiotics had surprising – and slightly worrying – consequences. ‘It increased the expression of genes that enabled the bacteria to take up DNA from their surroundings’, says Veening. This would facilitate the uptake of any resistant genes floating around.

What happened was that the antibiotic stalled the replication of the circular bacterial chromosome. This replication starts at a specific point, the origin of replication (oriC). When replication was stalled, the genes close to the oriC had already been copied. What is more, the cell attempted to replicate its chromosome afresh, and this again was stalled by the antibiotic, leading to even more copies of the genes near the oriC.
The genes facilitating the uptake of DNA turned out to be in this exact location. ‘The consequence is that multiple copies of these genes are present in the cell when replication is stalled’, explains Veening. ‘And extra copies mean increased production of the proteins encoded by these genes, like the ones that facilitate DNA uptake.’
Tip of the iceberg
This isn’t just odd, but looks suspiciously like a regulatory process: when bacteria are in trouble, they stimulate themselves to take up foreign DNA. ‘It is a very cheap way of regulating gene expression’, adds Slager. No proteins are involved, no regulatory cascades or anything: just location on the chromosome. Earlier this year, Veening and Slager and their co-workers published a second paper on genes that seem to benefit from their location. As a result, they were asked to write a review article on the importance of location.
When they began to dig deeper, their findings turned out to be the tip of the iceberg. They discovered, for example, that copying the chromosome sometimes takes longer than cell division. ‘In that case, a second replication cycle may start on the same chromosome.’ This will start from both the original oriC and the newly copied one. In this case, four copies of the genes near the starting point of replication will be present in the cell.
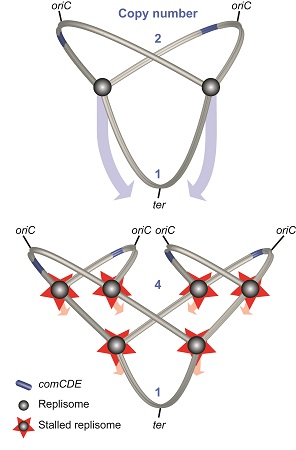
This difference in number of copies can affect the number of gene products, the proteins encoded by the genes. ‘But this is not always the case’, says Slager. ‘We found examples where the copy number doubled, and yet the number of gene products remained the same.’ So location is important, but the effect is not always straightforward. It is, however, one of the mechanisms to regulate genes. ‘Evolution is working on this’, says Veening. ‘All streptococcus species have these genes that facilitate DNA uptake near the origin of replication.’
Spores
Another example of the importance of location is seen in the Bacillus subtilis bacterium. In unfavourable conditions, it produces spores. To do this, the cell divides into a ‘mother cell’ and a ‘prespore’. The part that forms the prespore is smaller and initially contains only about 30 percent of the newly replicated chromosome. ‘The genes on this part are needed to produce a mature spore’, says Veening.
Other effects can be seen in some membrane proteins located on parts of the chromosome that are usually near the membrane. ‘Furthermore’, adds Slager, ‘proteins that work together in bacteria are usually encoded together on the chromosome, so they are produced in close proximity.’
Slager and Veening’s review is the first to summarize how bacteria regulate some genes according to their location on the chromosome, and should have quite some impact on microbiology. Slager says, ‘These location-driven differences in copy number may explain many findings already published in the literature.’
Reference: Jelle Slager and Jan-Willem Veening: Hard-Wired Control of Bacterial Processes by Chromosomal Gene Location. Trends in Microbiology, 27 June 2016 (corrected proofs) doi:10.1016/j.tim.2016.06.003
| Last modified: | 08 July 2016 10.08 a.m. |
More news
-
16 April 2024
UG signs Barcelona Declaration on Open Research Information
In a significant stride toward advancing responsible research assessment and open science, the University of Groningen has officially signed the Barcelona Declaration on Open Research Information.
-
02 April 2024
Flying on wood dust
Every two weeks, UG Makers puts the spotlight on a researcher who has created something tangible, ranging from homemade measuring equipment for academic research to small or larger products that can change our daily lives. That is how UG...
-
18 March 2024
VentureLab North helps researchers to develop succesful startups
It has happened to many researchers. While working, you suddenly ask yourself: would this not be incredibly useful for people outside of my own research discipline? There are many ways to share the results of your research. For example, think of a...

