Projects
PHOTOSYSTEM I
Photosystem I (PSI) is a multi-subunit protein present in green plants and cyanobacteria. For green plants, we are interested in the structural determination of the peripheral membrane-bound subunits, which are not present in cyanobacteria. Since the PSI structure of cyanobacteria has been solved by X-ray diffraction, only interactions with other peripheral proteins remain of interest for electron microscopical studies. Here , you will find a description of the structural determination of a giant complex between PSI and Isia, which appeared in Nature, August 2001.
PHOTOSYSTEM II COMPLEX FROM ARABIDOPSIS
The Photosystem II (PS II) complex is the abundant large membrane protein in the stacked parts of the thylakoid membrane of green plant chloroplasts. The complex from green plants consists of about 25 different subunits, of which one half is directly involved in light-harvesting. Over the years we have studied various types of PSII preparations from spinach.
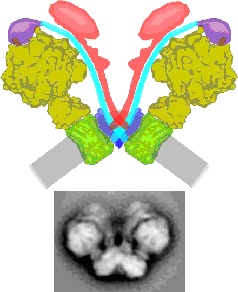
RESPIRATORY CHAIN SUPERCOMPLEXES IN MITOCHONDRIAL MEMBRANES
Mitochondria have an intricate heavily folded inner membrane which houses the respiratory chain complexes which are, together with the ATP synthase complex, responsible for energy production stored as ATP. The structure of the individual membrane-bound protein components have been well characterized, especially by X-ray diffraction studies. During the past years increasing evidence that these components are organized into supercomplexes was obtained, particularly by the application of Blue-native polyacrylamide gel electrophoresis. We use single particle electron microscopy for a structural characterization of some of these supercomplexes. Up till now we characterized the supercomplex composed of monomeric Complex I and dimeric Complex III from Arabidopsis (dutch: Zandraket) and more recently the dimeric complex of ATP synthase from Polytomella (a colorless alga related to Chlamydomonas). The ATP synthase is substantially kinked in its membrane-embedded domains and this allows also for the first time to define a functional role for these supercomplexes. The dimeric ATP synthase complex appears to be responsible for the folding of the inner mitochondrial membrane.
AUTOMATION OF EM DATA ACQUISITION

A system has been developed for semi-automatic specimen selection and data acquisition for protein electron crystallography, based on a slow-scan CCD camera connected to a transmission electron microscope and control from an external computer. Areas of interest on the specimen are localised at low magnification and subsequently imaged on the CCD camera, using a dose which is small compared to the dose used in the exposure mode. The crystalline quality of the area is evaluated from the appearance of diffraction peaks in the calculated image Fourier transform. If the quality is considered good, images can then be recorded in different modes, both on film and using the CCD camera. Using this system a significant gain, both quantitatively and qualitatively, can be obtained in acquiring data for electron crystallography of beam-sensitive materials.
A freely available image processing package GRIP (Groningen image processing), has been developed, which is aimed at 2D image analysis of 2D protein crystals and single particle analysis. Routines for use on clusters of computers have been and are still being developed, using LAM/MPI parallel computing libraries, to increase processing speed of large data sets. Images made with other packages and on machines with other hardware architecture than the target machine are automatically converted.
The program can also be used as an easy to use interface to the 3D image analysis programs from the MRC labs in Cambridge. It adds display functionality and routines for lattice refinement, origin refinement and symmetrisation of structure factors.
The Grip image format is compatible with the IMAGIC image format. Going from 2D to 3D (for single particles) the EMAN package fits seamless together with Grip.
ELECTRON TOMOGRAPHY
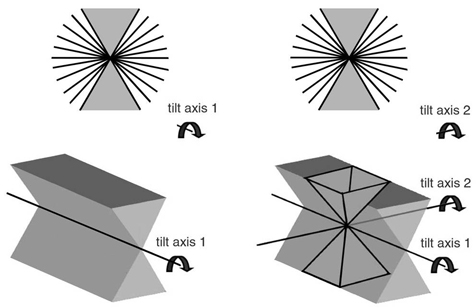
Electron tomography is a method to reconstruct the 3D volume of large objects, such as organelles and large protein complexes from series of tilted images. The method is very attractive to study large protein complexes in reconstructed organelles, such as mitochondria and chloroplasts. Subvolumes from tomograms of these organelles containing ATP synthase or photosystem II can be processed by 3D averaging. This provides novel insight how these complexes are arranged in these organelles. Best results are obtained when tomography is performed as dual-axis electron tomography: First, one tilt series is performed, then the specimen is rotated by 90° and then a second tilt series is recorded. Combining these will limit the 3D volume that can not be sampled (because the tilt holder can not be tilted more than about 60°) from a missing wedge to a missing pyramid.
See our papers:
Dudkina, Oostergetel, Braun and Boekema (2010) Row-like organization of ATP synthase in intact mitochondria determined by cryo-electron tomography Biochim. Biophys. Acta 1797, 272–277
Dudkina, Kouřil, Bultema, Boekema, EJ (2010) Imaging of organelles by electron microscopy reveals protein–protein interactions in mitochondria and chloroplasts FEBS Lett. 584, 2510–2515
Kouřil, Oostergetel and Boekema (2011) Fine structure of granal thylakoid membrane organization using cryo electron tomography Biochim. Biophys. Acta 1807, 368–374
| Last modified: | 03 October 2012 11.40 a.m. |
