New insights into antibiotic resistance development
We know little about how microbes develop resistance to antibiotics. We do know that resistance mutations occur in individual cells, but nearly all the work on antibiotic resistance uses large numbers of bacteria grown in cultures or as colonies on plates. What is lacking, according to microbiologists Robin Sorg and Jan-Willem Veening from the University of Groningen, is a detailed look at the single cell. They published their study of the prerequisites for antibiotic resistance development in the important human pathogen Streptococcus pneumoniae on 30 October in the journal Nature Communications.
The basic picture is clear. When you expose millions of bacteria to a low dose of an antibiotic, a few cells may survive. Eventually, they may even become fully resistant and start replicating. A neat picture, but not very detailed.
‘These types of studies are usually done with cell cultures, where the growth of bacteria is determined by measuring the optical density of nutrient broth’, explains Robin Sorg. ‘But this only gives you limited information on how the resistant bacterium is able to develop and outcompete the others. Antibiotic resistance comes at a cost, so resistance only becomes prevalent when cells have an advantage.’ He and fellow microbiologist Jan-Willem Veening therefore studied the effect of antibiotics on bacteria growing under a microscope. The results provide an important insight into the development of antibiotic resistance.
Key to understanding
Sorg, who will defend his PhD thesis under Veening’s supervision sometime next year, studied the human pathogen Streptococcus pneumoniae (pneumococcus). He observed how it grew when exposed to different antibiotics. ‘Some single-cell studies have been done – on the way antibiotics affect bacteria – but none of these systematically analysed growth kinetics, metabolic activity and cell-to-cell-variability. However, these factors could be key to understanding and predicting resistance development’, he says.
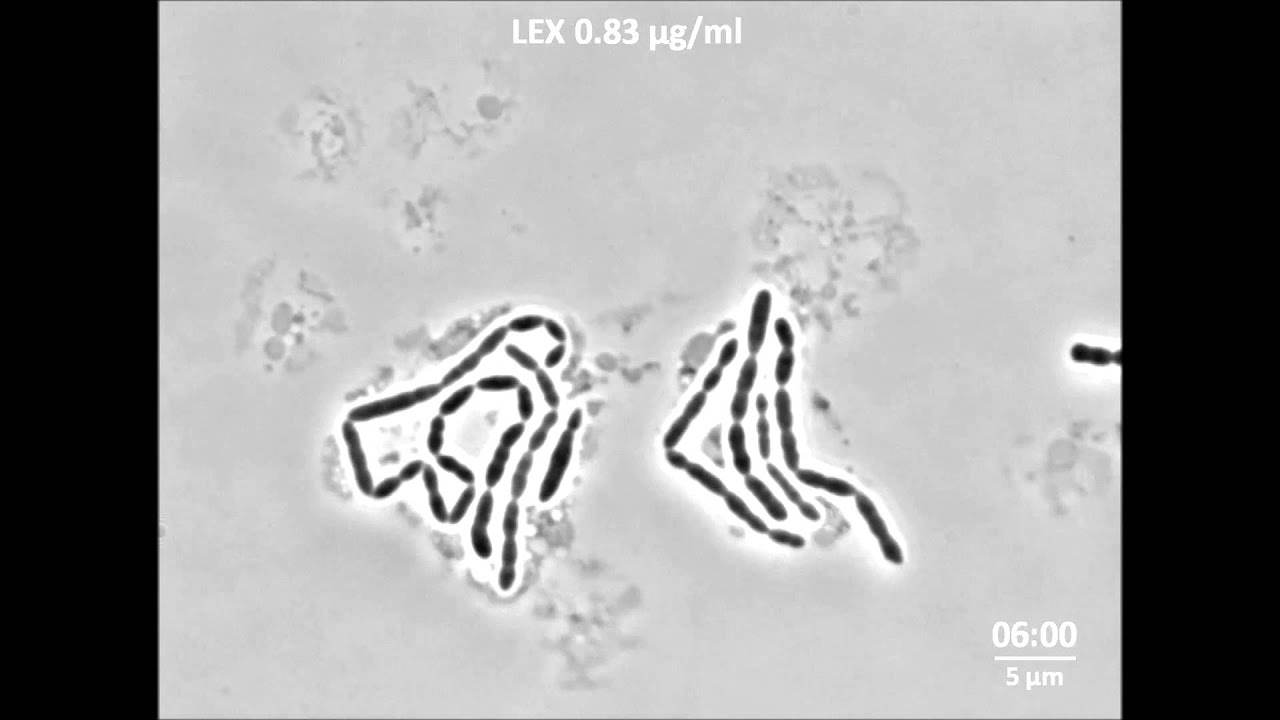
He started his experiments with just a few cells in a special incubation chamber on a microscope. Time lapse imaging showed the growth of these bacteria – or lack thereof. He then used different dosages of antibiotics that either directly kill the bacteria or stop their growth, and measured the growth kinetics. He also used a special bioluminescence marker to measure gene expression inside the cells, which indicates how healthy and active the bacteria are.
When exposed to a bacteriostatic antibiotic, the cells were promptly affected, and at a higher concentration they grew more slowly. ‘But the morphology of the cells had changed, they looked seriously perturbed.’ After a number of hours, the cells eventually stopped growing. ‘We noticed striking differences in antibiotic sensitivity between cells of identical genetic origin’, explains Sorg. ‘This suggests that epigenetic factors play a role in determining when individual cells experience selection pressure towards resistance development.’
Mutant selection
They repeated the experiments, but this time with a bactericidal antibiotic. ‘At first, the bacteria grew at normal rates, whatever the concentration, but then their growth rate slowed down and they died. So the cell damage caused by this type of antibiotic does not instantaneously affect growth, and it is worth noting that it is always driven by cellular metabolic processes.’ These cells looked surprisingly normal until the end.

In all the experiments, cells that stopped growing could still be active, with a functioning metabolism and gene expression. ‘That is important for the development of resistance’, says Sorg. His data indicates that in most cases a non-genetic mechanism ensures cells survive the initial onslaught of antibiotics. Mutations that confer resistance appear spontaneously later in the process. ‘The cells need time to acquire these mutations, so their initial survival is crucial.’ The time between administration of the antibiotic that impacts bacterial growth and cell death is therefore called the ‘mutant selection window’. This is vital in understanding the genesis of antibiotic resistance.
‘When cells die slowly, there is more time for genetic resistance to kick in. And some antibiotics may even stimulate mutations.’ But as cellular metabolic processes decrease, so do does the likelihood of new mutations. This is important information for drug development, Sorg stresses: ‘Drugs that quickly reduce cell activity will make it less likely that the bacteria become resistant.’
The work also shows that drug dosage is important. ‘Once a patient has taken an antibiotic, its concentration may vary between tissues and over time. To minimize the development of antibiotic resistance, it is probably better to prescribe a higher dose than is strictly required to cure the patient. That would increase the chance of side effects, so it is a trade-off between the interest of the individual patient and that of future patients relying on effective antibiotics for which resistance has not spread.’
Reference: Robin A. Sorg and Jan-Willem Veening, Micro-scale insights into pneumococcal antibiotic mutant selection windows , Nature Communications 30 oktober, DOI 10.1038/NCOMMS9773
| Last modified: | 03 November 2017 2.30 p.m. |
More news
-
15 April 2024
Single-molecule engineering niche in Gravitation research
With her expertise in single-molecule techniques, Dr. Kasia Tych (GBB) will contribute to a big Grvitation-research programme
-
15 April 2024
Night vision with artificial atoms
Every two weeks, UG Makers puts the spotlight on a researcher who has created something tangible, ranging from homemade measuring equipment for academic research to small or larger products that can change our daily lives. That is how UG...
-
09 April 2024
University of Groningen to become member of the 4TU Stan Ackermans Institute
Through this membership, the two EngD programmes in Autonomous Systems and in Sustainable Process Design at the Faculty of Science and Engineering will be added to this partnership with the 4TU from September 2024
